Very excited to share this work by the heroic team - @carolilucas Pat Wong @sneakyvirus @tiago21bio et al on “Longitudinal analyses reveal immunological misfiring in severe COVID-19”. A thread on our findings. #COVID19 #Pathogenesis (1/n) https://www.nature.com/articles/s41586-020-2588-y">https://www.nature.com/articles/...
In this study, we enrolled COVID+ patients and analyzed their immune responses and viral load over the course of their hospital stay. We also compared samples from COVID- healthy health care workers (HCWs) as comparison. (2/n)
As reported by others, we found that COVID patients with severe disease have low T cell numbers and increased monocytes and neutrophils. We also found eosinophils come up in patients. This is bizarre  https://abs.twimg.com/emoji/v2/... draggable="false" alt="🧐" title="Gesicht mit Monokel" aria-label="Emoji: Gesicht mit Monokel"> (3/n)
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🧐" title="Gesicht mit Monokel" aria-label="Emoji: Gesicht mit Monokel"> (3/n)
The serum cytokines indicate the activation of inflammasomes based on elevated cytokines IL-18 and IL-1b, as well as IL-6. For more on the #Inflammasome and #COVID19, see review by @jeremymmunology @MiyuMoriyama (4/n) https://www.jimmunol.org/content/early/2020/06/02/jimmunol.2000513">https://www.jimmunol.org/content/e...
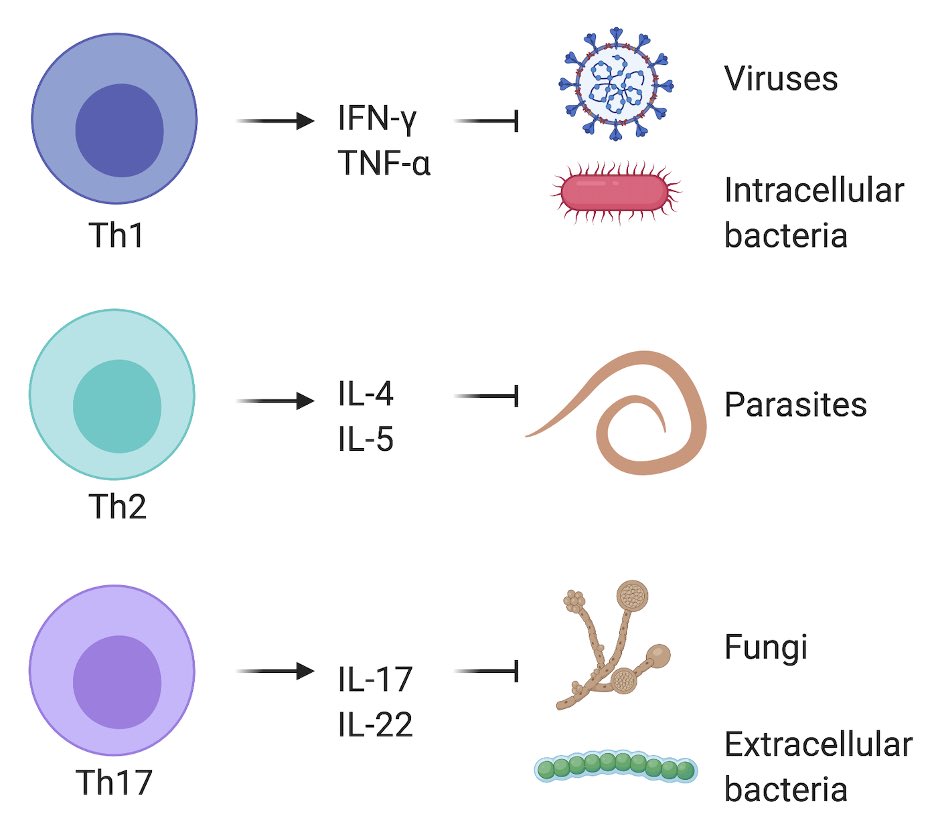
The immune system makes responses that are best suited for different types of pathogens. For example, different flavors of CD4 T cells are made to combat viruses/intracellular bacteria (Th1), fungi/extracellular bacteria (Th17) and worms (Th2) infection. (5/n)
In severe #COVID19, we find all cytokine types being elevated overtime in patients. Even eosinophils and IgE which are good for expelling worms and not for viral defence became elevated in severe cases. (6/n)
So, what is driving these prolonged immune responses in severe #COVID19? One clue comes from this figure, where we found that the nasal viral load fails to come down over time in patients with severe disease. Data generated with @NathanGrubaugh @awyllie13 teams  https://abs.twimg.com/emoji/v2/... draggable="false" alt="💪🏼" title="Angespannter Bizeps (mittelheller Hautton)" aria-label="Emoji: Angespannter Bizeps (mittelheller Hautton)"> (7/n)
https://abs.twimg.com/emoji/v2/... draggable="false" alt="💪🏼" title="Angespannter Bizeps (mittelheller Hautton)" aria-label="Emoji: Angespannter Bizeps (mittelheller Hautton)"> (7/n)
Is it that patients with severe disease fail to produce antiviral interferons? No! They are making more interferons and other innate cytokines in response to viral load, suggesting that these #IFNs are not able to control virus in patients. (8/n)
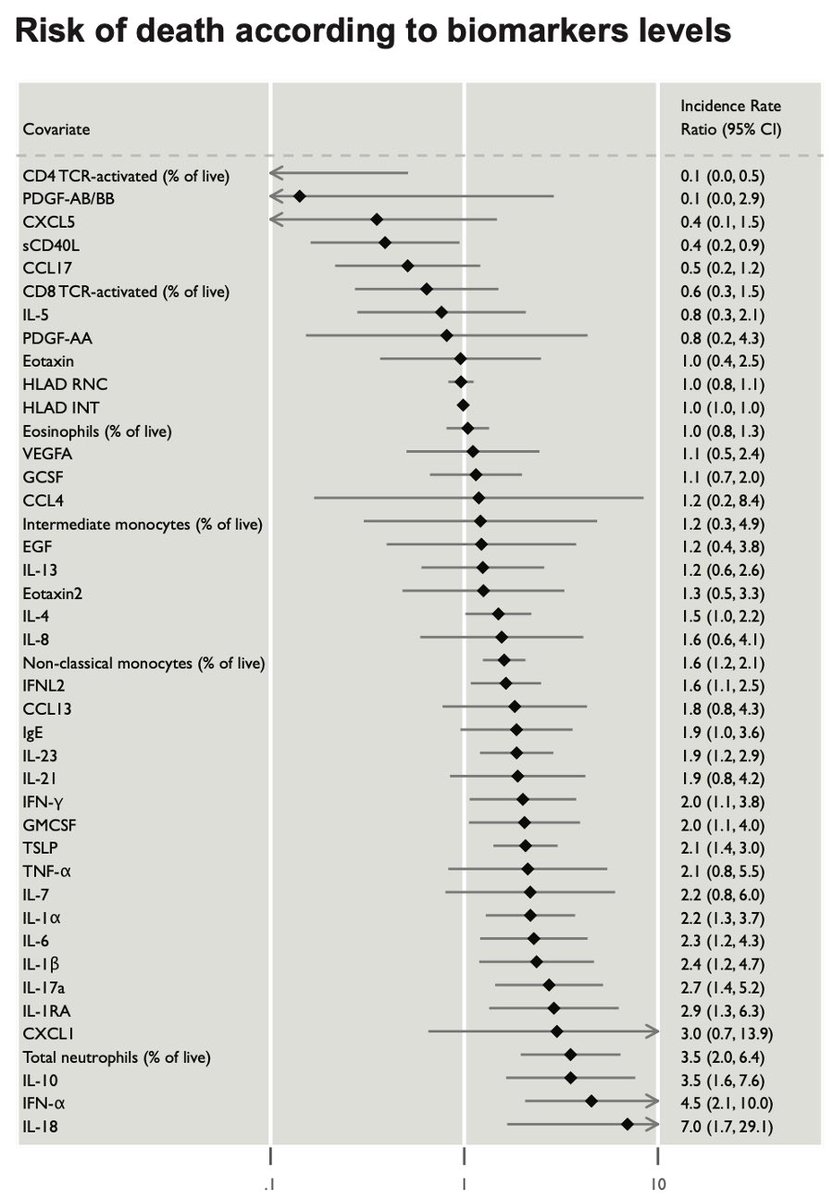
Also, elevated levels of IFN-a and IL-1RA within the first 12 days of symptom correlates with mortality and with longer hospital stay. (9/n)
These data build nicely on others’ seminal work @Lo_Zanzi and Peng Hong showing the detrimental effects of late IFNs in #COVID19. (10/n)
https://science.sciencemag.org/content/early/2020/06/10/science.abc3545
https://science.sciencemag.org/content/e... href=" https://www.cell.com/cell-host-microbe/fulltext/S1931-3128(20)30401-7">https://www.cell.com/cell-host...
https://science.sciencemag.org/content/early/2020/06/10/science.abc3545
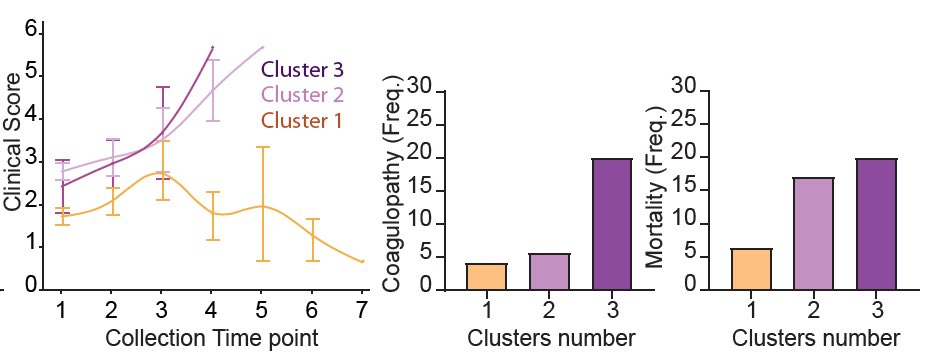
We next used unsupervised clustering to see if patients fall into different groups based on their cytokine levels within the first 12 days of symptom. This revealed 3 clusters of patients based on 4 immune signatures. (11/n)
We then asked if we take all patients and all time points and group them based on their cytokine profile, what do we get? We got a remarkably similar clustering based on almost identical immune signatures. (12/n)
Moreover, we saw distinct disease trajectories for the three clusters of patients. Patients in cluster 1 enriched in tissue repair growth factors mostly recovered. Those in cluster 2 and 3, enriched in chemokines and mixture of cytokines did worse. (13/n)
How can we use these insights for future treatment? First, we found biomarkers for mortality. An amazing work by @mariasundaram @Muhellingson @SaadOmer3. These biomarkers can be a useful prognostic tool for better targeted therapy. (14/n)
Also, our trajectory analyses raise the possibility that early interventions that target inflammatory markers that are predictive of worse disease outcome would be more beneficial than targeting late-appearing cytokines.  https://abs.twimg.com/emoji/v2/... draggable="false" alt="👀" title="Augen" aria-label="Emoji: Augen"> at you #inflammasomes (15/n)
https://abs.twimg.com/emoji/v2/... draggable="false" alt="👀" title="Augen" aria-label="Emoji: Augen"> at you #inflammasomes (15/n)
Lastly, this work is a huge collaboration across @YaleIBIO @YaleMed @YNHH @YaleSPH @YaleIDFellows @YaleCancer @YaleNursing @YalePCCSM @YaleGH @yale_Labmed @RockefellerUniv @HHMINEWS. Enormously rewarding to learn from our patients -my first translational immunology paper  https://abs.twimg.com/emoji/v2/... draggable="false" alt="🙏🏼" title="Folded hands (mittelheller Hautton)" aria-label="Emoji: Folded hands (mittelheller Hautton)"> (end)
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🙏🏼" title="Folded hands (mittelheller Hautton)" aria-label="Emoji: Folded hands (mittelheller Hautton)"> (end)

 Read on Twitter
Read on Twitter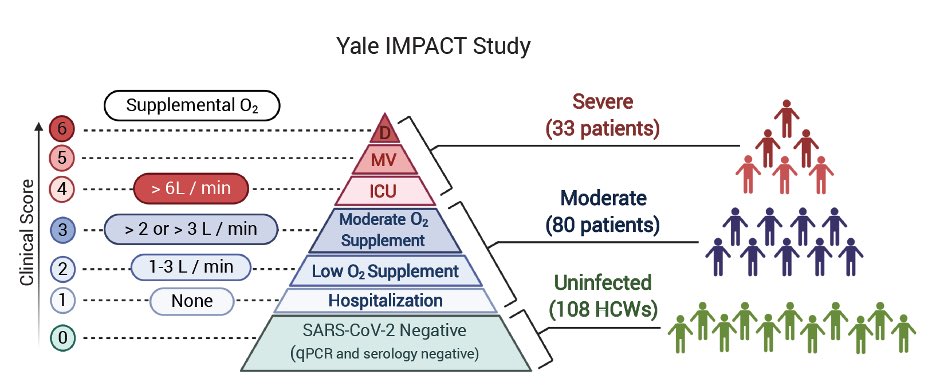
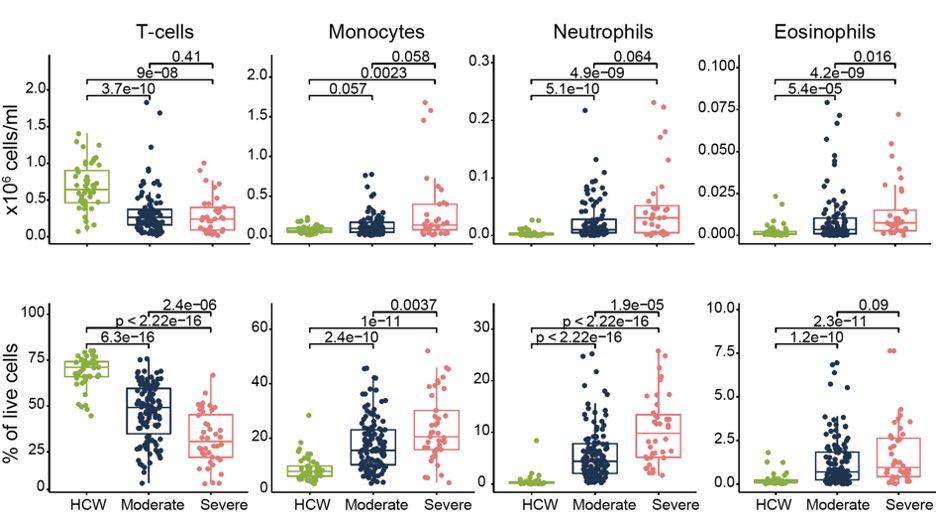 (3/n)" title="As reported by others, we found that COVID patients with severe disease have low T cell numbers and increased monocytes and neutrophils. We also found eosinophils come up in patients. This is bizarre https://abs.twimg.com/emoji/v2/... draggable="false" alt="🧐" title="Gesicht mit Monokel" aria-label="Emoji: Gesicht mit Monokel"> (3/n)" class="img-responsive" style="max-width:100%;"/>
(3/n)" title="As reported by others, we found that COVID patients with severe disease have low T cell numbers and increased monocytes and neutrophils. We also found eosinophils come up in patients. This is bizarre https://abs.twimg.com/emoji/v2/... draggable="false" alt="🧐" title="Gesicht mit Monokel" aria-label="Emoji: Gesicht mit Monokel"> (3/n)" class="img-responsive" style="max-width:100%;"/>
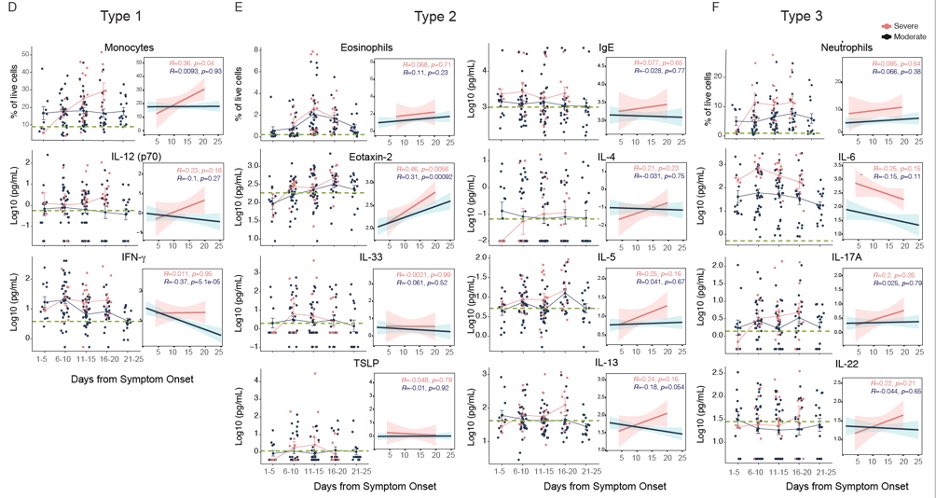
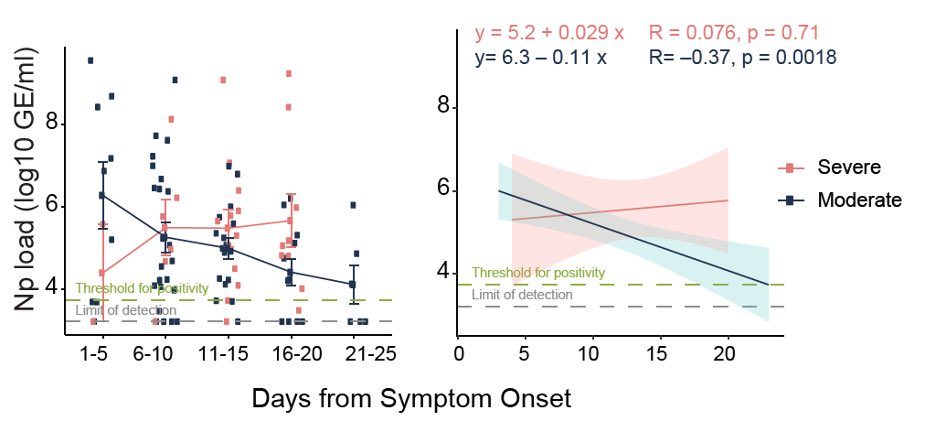 (7/n)" title="So, what is driving these prolonged immune responses in severe #COVID19? One clue comes from this figure, where we found that the nasal viral load fails to come down over time in patients with severe disease. Data generated with @NathanGrubaugh @awyllie13 teams https://abs.twimg.com/emoji/v2/... draggable="false" alt="💪🏼" title="Angespannter Bizeps (mittelheller Hautton)" aria-label="Emoji: Angespannter Bizeps (mittelheller Hautton)"> (7/n)" class="img-responsive" style="max-width:100%;"/>
(7/n)" title="So, what is driving these prolonged immune responses in severe #COVID19? One clue comes from this figure, where we found that the nasal viral load fails to come down over time in patients with severe disease. Data generated with @NathanGrubaugh @awyllie13 teams https://abs.twimg.com/emoji/v2/... draggable="false" alt="💪🏼" title="Angespannter Bizeps (mittelheller Hautton)" aria-label="Emoji: Angespannter Bizeps (mittelheller Hautton)"> (7/n)" class="img-responsive" style="max-width:100%;"/>
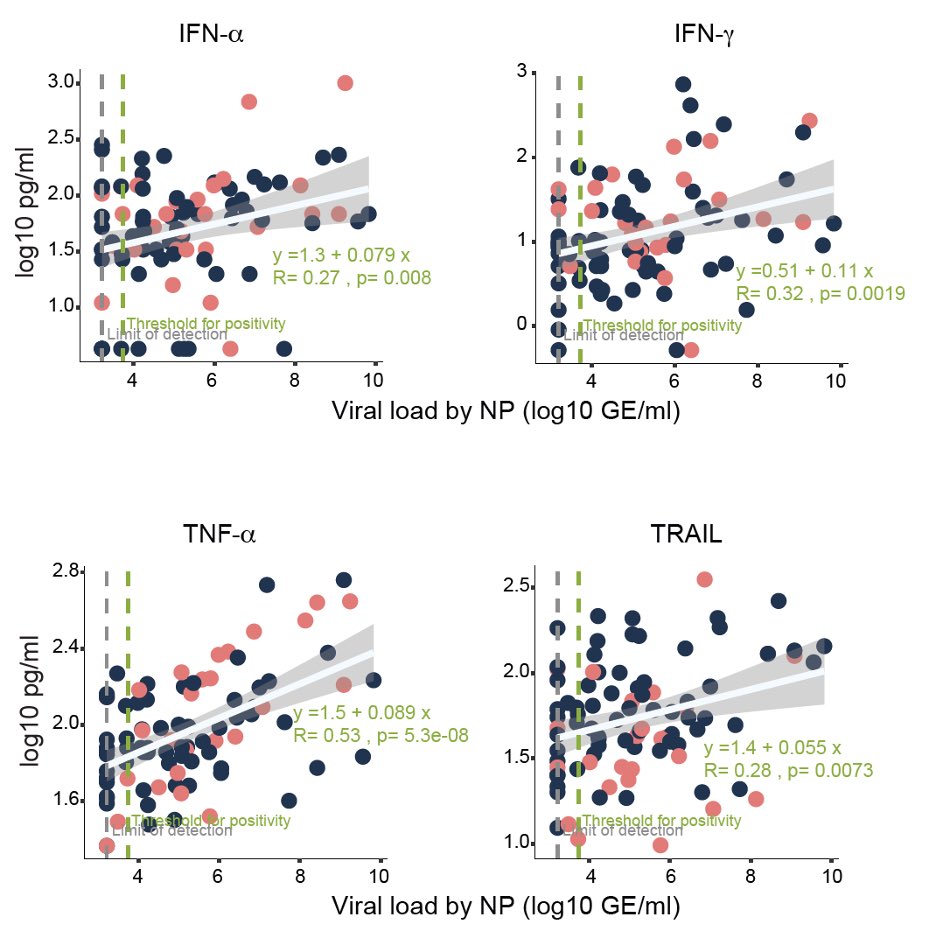
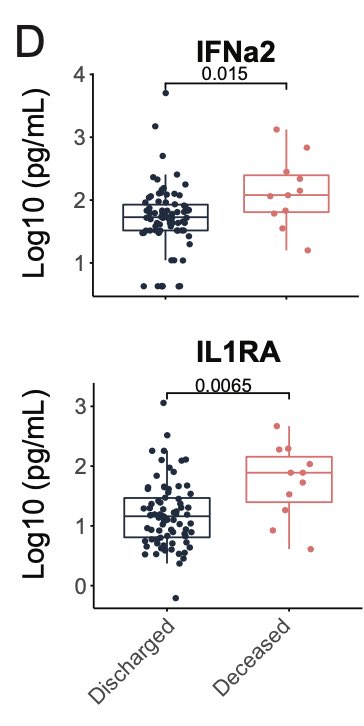

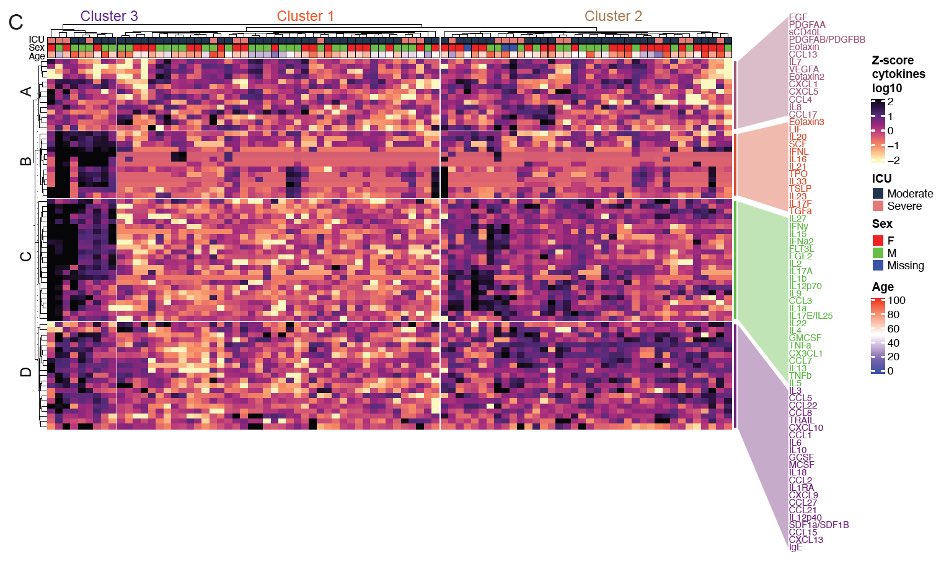

 (end)" title="Lastly, this work is a huge collaboration across @YaleIBIO @YaleMed @YNHH @YaleSPH @YaleIDFellows @YaleCancer @YaleNursing @YalePCCSM @YaleGH @yale_Labmed @RockefellerUniv @HHMINEWS. Enormously rewarding to learn from our patients -my first translational immunology paper https://abs.twimg.com/emoji/v2/... draggable="false" alt="🙏🏼" title="Folded hands (mittelheller Hautton)" aria-label="Emoji: Folded hands (mittelheller Hautton)"> (end)" class="img-responsive" style="max-width:100%;"/>
(end)" title="Lastly, this work is a huge collaboration across @YaleIBIO @YaleMed @YNHH @YaleSPH @YaleIDFellows @YaleCancer @YaleNursing @YalePCCSM @YaleGH @yale_Labmed @RockefellerUniv @HHMINEWS. Enormously rewarding to learn from our patients -my first translational immunology paper https://abs.twimg.com/emoji/v2/... draggable="false" alt="🙏🏼" title="Folded hands (mittelheller Hautton)" aria-label="Emoji: Folded hands (mittelheller Hautton)"> (end)" class="img-responsive" style="max-width:100%;"/>


