There are many problems with Difference-in differences design, but many of them can be addressed (or evaluated more clearly) If you follow a rule of 1) stack by event date, 2) insist on "clean controls."
Burgeoning literature on how staggered or on-and-off treatments (along with het. treatment effects) can cause problems - due to negative weights. @CdeChaisemartin @borusyak @XJaravel @agoodmanbacon and many others.
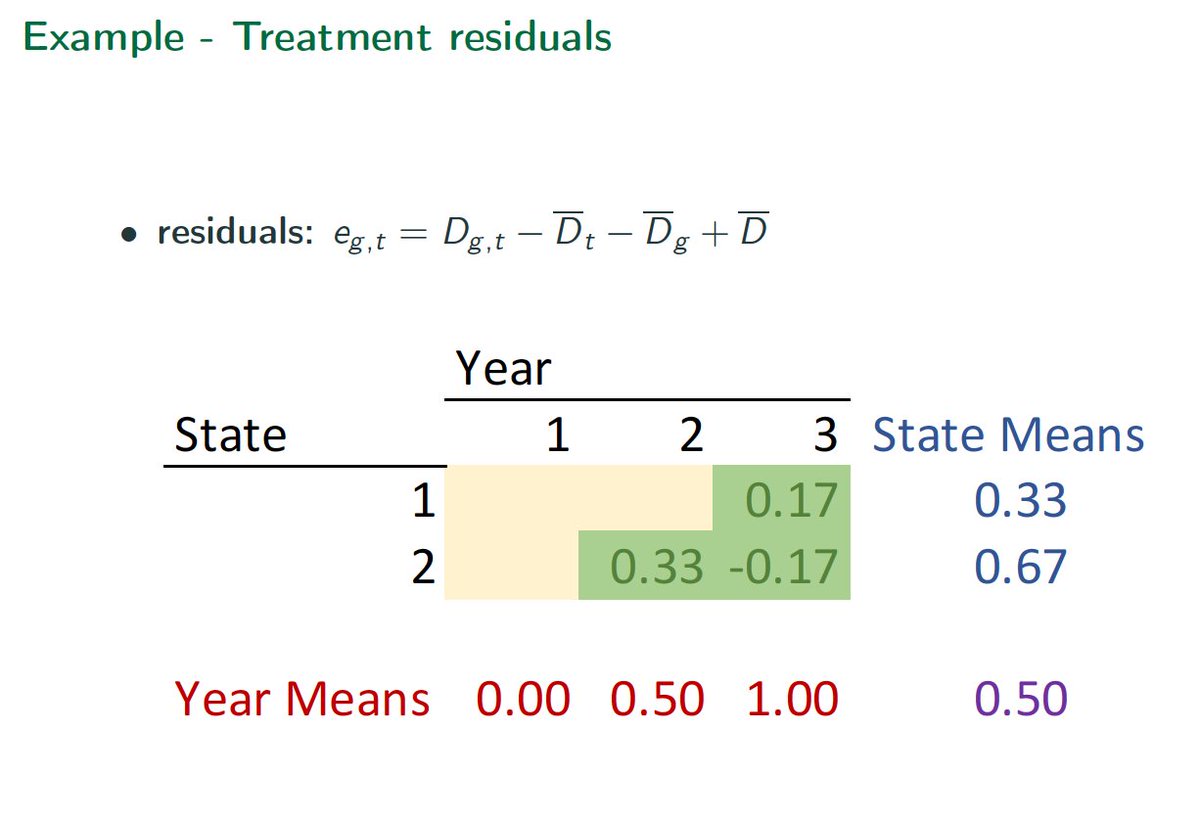
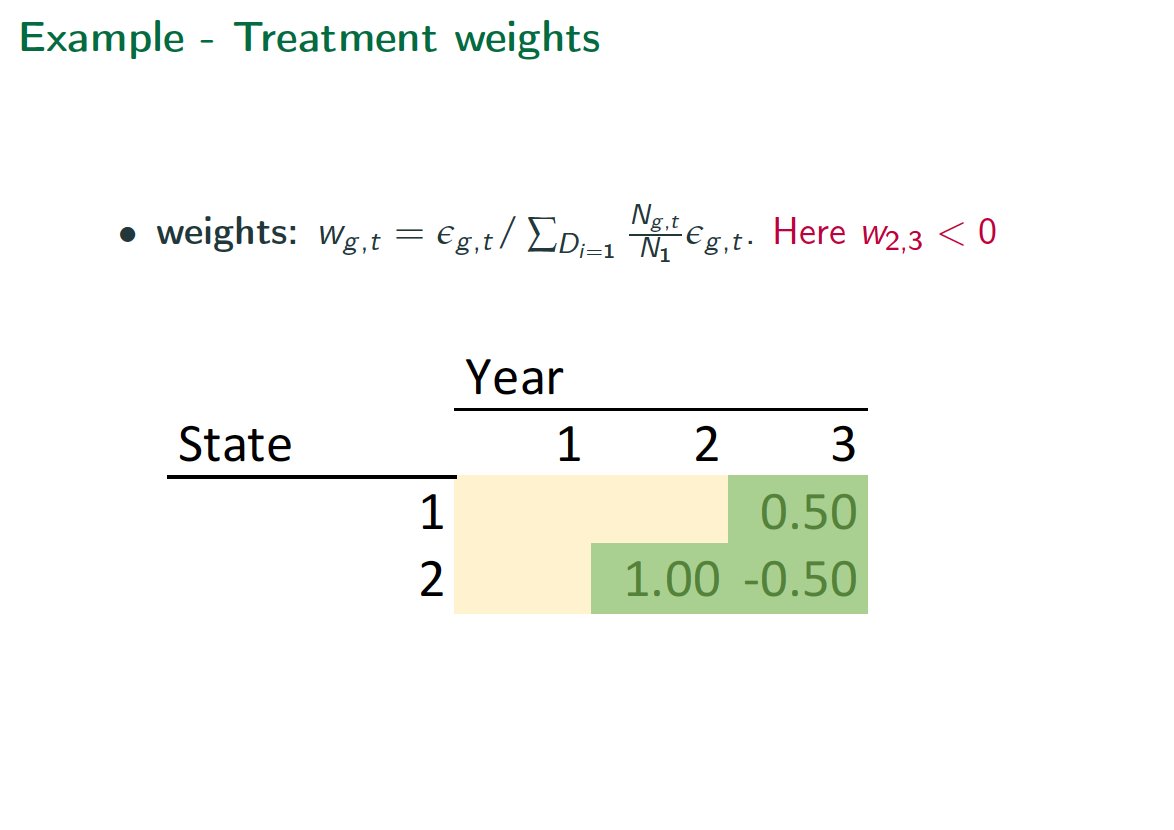
What& #39;s the problem? Easiest to see with staggered design. Later treatments in early adopters tend to get negative weights.
Bad.
Bad.
Solution is quite simple conceptually. Use clean controls for each treatment event. Many different ways of doing this, but conceptually very similar.
Here is de Chaisemartin & d& #39;Haultfoeuille DID_m estimator: take every treatment, find only clean control, do DID, and average.
Here is de Chaisemartin & d& #39;Haultfoeuille DID_m estimator: take every treatment, find only clean control, do DID, and average.
Interestingly, this is closely related to what we did in our QJE 2019 paper using a "stacked design." Same idea.Take every treatment, find all clean controls, and do DID. And average. Slightly different in details of implementation.
We stacked by event date to get averaged effect by event date. But deC and dH can do similarly by taking longer differences, as their code shows. So very similar!
Similarly, interactive weight estimator of Abraham and Sun is conceptually quite similar. Dummy out all post-treatment periods in every treated unit. This prevents early adopters to be controls. Then average them. Conceptually very, very similar.
I.e., just use clean controls!
I.e., just use clean controls!
I can go on, but the main point is this. DID with staggered or otherwise time-varying treatment can create problems because a simple TWFE reg doesn& #39;t factor in who is a clean control. But you can do that. And there are many, similar ways of doing that.
Final word on why I like the "stacked approach."
Take every treated unit. Find all clean controls. Call it an "experiment." Stack by all "experiments" sorted by event time. Only use within-"experiment" variation by using "expieriment x calendar time effects.) Pool across events
Take every treated unit. Find all clean controls. Call it an "experiment." Stack by all "experiments" sorted by event time. Only use within-"experiment" variation by using "expieriment x calendar time effects.) Pool across events
So why do I like this? Because it allows us not just to deal with the negative weighting issue but be super clear about where identification is coming from - for every experiment! Maybe some experiments fail some falsification. Then we drop those, or regularize in some way. Etc.
For more on a relatively straightforward example of pooled, stacked event studies, see Appendix G of our 2019 QJE paper here: https://academic.oup.com/qje/article/134/3/1405/5484905">https://academic.oup.com/qje/artic...
Here are links to my slides for DID, TWFE lectures. Doing them last week and this week for our grad applied micro class! https://www.dropbox.com/s/kyrbudhnp43g8xc/Lecture_7_8_9_FE_DID_and_Inference_-_2020_wpause.pdf?dl=0">https://www.dropbox.com/s/kyrbudh...

 Read on Twitter
Read on Twitter