On Feb 29, using the genetic sequence of the first community case of #COVID19 in Washington State, I made the claim that the Washington State outbreak descended directly from the Jan 15 arrival of the WA1 case with direct travel history to Wuhan. 1/18 https://twitter.com/trvrb/status/1233970271318503426">https://twitter.com/trvrb/sta...
Based on data that& #39;s emerged in the intervening months, I no longer believe that a direct WA1 introduction is a likely hypothesis for the origin of the Washington State outbreak. 2/18
The central observation had been that the Washington State outbreak genetically matched the WA1 virus, but possessed an additional two mutations as expected given sampling times. 3/18
This results in the following tree structure, with WA1 directly ancestral to the Washington State outbreak (in red). 4/18
However, given the rate at which SARS-CoV-2 accumulates genetic differences, it& #39;s possible to have this tree structure even if WA1 sits on a side branch relative to the Washington State outbreak. 5/18
I had initially thought this unlikely given that this "side branch" scenario would mean that the genetic match between WA1 and the Washington State outbreak was strictly by chance, while this variant is rare in viruses from China (found in ~3% of sequenced viruses). 6/18
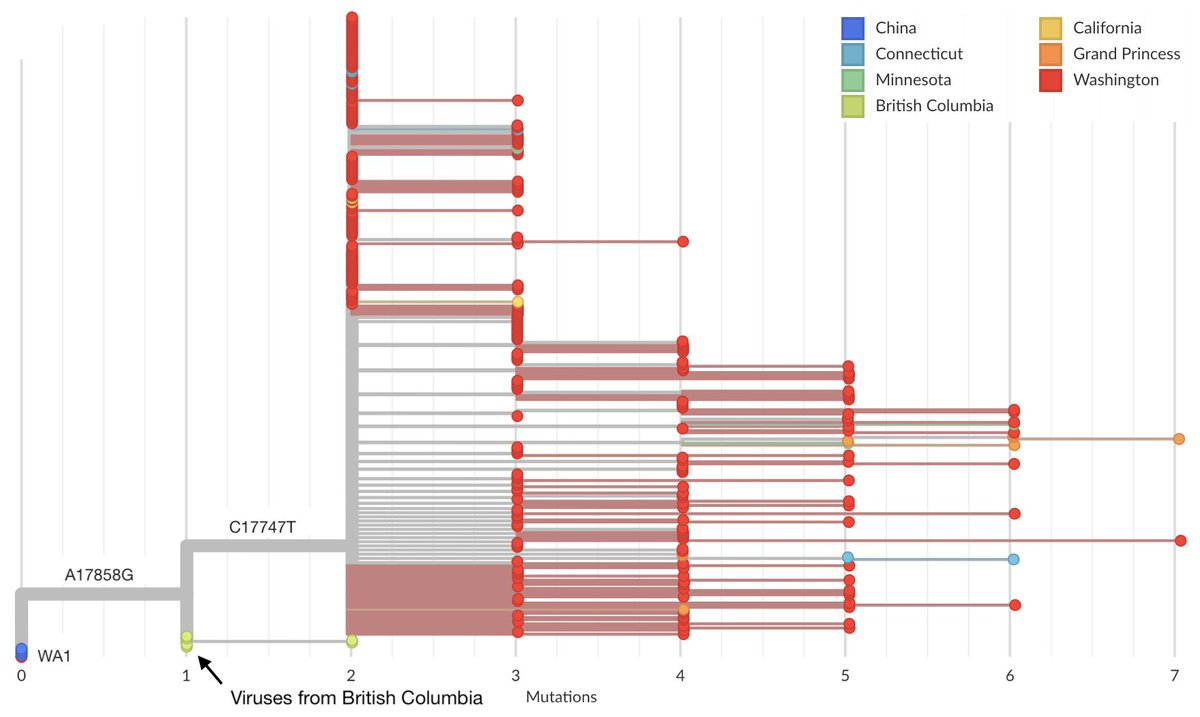
However, additional viruses from British Columbia sequenced by @CDCofBC and shared March 31 interdigitate between WA1 and the Washington State outbreak clade. 7/18
These viruses were sampled from British Columbia between March 5 and 12, but outgroup relative to WA viruses, possessing the 17858G mutation but not the additional 17747T mutation possessed by the Washington State outbreak clade. 8/18
A scenario of a WA1 introduction requires a transmission chain that makes it to British Columbia while not accumulating mutations and a local transmission chain that accumulates two mutations. 9/18
The observation that all viruses in the Washington State outbreak clade differ by two mutations from WA1 is important. I had initially expected this two mutation "gap" to be filled in with sequencing additional viruses . 10/18
However, with 277 genomes sequenced from WA viruses belonging to the clade collected between Feb 21 and March 15 we don& #39;t see any samples identical to WA1 or just possessing 17858G. 11/18
In this preprint, Mike Worobey, @LemeyLab and colleagues run phylogenetic outbreak simulations to demonstrate that this two mutation gap is not statistically likely with a WA1 introduction. https://www.biorxiv.org/content/10.1101/2020.05.21.109322v1">https://www.biorxiv.org/content/1... 12/18
At this point, I don& #39;t think we can exclude a WA1 introduction as an impossibility, but I think the most likely scenario is a separate undetected introduction by a genetically identical virus or a virus possessing the additional 17858G mutation. 13/18
This separate introduction may have been to British Columbia or may have been elsewhere. Better resolving this introduction geographically would benefit from additional sequencing of samples collected closer in time to the introduction event. 14/18
My best estimate for the date of the introduction of the Washington State outbreak clade derives from a molecular clock analysis using just viruses sampled from this clade in Washington State. https://www.medrxiv.org/content/10.1101/2020.04.02.20051417v2">https://www.medrxiv.org/content/1... 15/18
Doing so gives an estimated common ancestor between Jan 18 and Feb 9, or between 3 and 6 weeks before community transmission was detected on Feb 28. The introduction would likely have occurred slightly before this common ancestor virus existed. 16/18
Thus, I believe I was wrong in the original assessment of a WA1 introduction, but correct in asserting significant community spread in Washington State on Feb 29. 17/18
I& #39;m sorry to have created confusion here. Although I do think that my original actions were warranted given available evidence at the time. 18/18

 Read on Twitter
Read on Twitter