I have been immersed in the concept of ‘non-genetic individuality’ for the past few months while writing/revising papers and wanted to do a thread summarizing the field as I think it may be interesting for some! So here goes (it& #39;s LONG!): (1/27)
Non-genetic individuality merely refers to the phenotypic variation observed in a population of individuals expected to have ‘identical’ genomes. Of course, that is often not explicitly demonstrated and will be discussed here.
The concept arises first in research focused on understanding how phenotypic variability arises in single celled organisms like bacteria, and how this variability can influence evolution of isogenic (same genomes) colonies of bacteria.
Using state of the art single cell methods to catalogue ‘straight swimming’ or ‘tumbling’ behaviors – Spudich and Koshland monitored whether individuals had variable capacity to initiate ‘tumbling’ under homogenous conditions.
https://www.nature.com/articles/262467a0">https://www.nature.com/articles/... https://www.youtube.com/watch?v=IOl6q9P3xAg">https://www.youtube.com/watch...
https://www.nature.com/articles/262467a0">https://www.nature.com/articles/... https://www.youtube.com/watch?v=IOl6q9P3xAg">https://www.youtube.com/watch...
They observed high degrees of interindividual variability which persisted throughout the lifespan of an individual bacteria – and posited that these differences may arise as a result of uneven partitioning of cytoplasmic components during division (asymmetric division).
Later (skipping over key studies for time’s sake) the ability to monitor the transcriptional kinetics of individual genes over time in bacteria revealed that stochasticity in gene expression could be attributed to noisy ‘bursts’ of transcription.
https://pubmed.ncbi.nlm.nih.gov/12183631/ ">https://pubmed.ncbi.nlm.nih.gov/12183631/...
https://pubmed.ncbi.nlm.nih.gov/12183631/ ">https://pubmed.ncbi.nlm.nih.gov/12183631/...
Such bursts lead to fluctuations in protein expression manifesting in a range of protein expression levels for a population of cells – if this seems abstract, I invite you to think about the variation in cell types that you may be taken for granted in your everyday analysis:
The early 2000s had many papers on bacteria and yeast which explored the source and of biological noise in single cells. Here are a few interesting papers:
https://pubmed.ncbi.nlm.nih.gov/16699522/
https://pubmed.ncbi.nlm.nih.gov/16699522/... href=" https://pubmed.ncbi.nlm.nih.gov/15790857/ ">https://pubmed.ncbi.nlm.nih.gov/15790857/... https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1410811/">https://www.ncbi.nlm.nih.gov/pmc/artic...
https://pubmed.ncbi.nlm.nih.gov/16699522/
These papers tried to isolate individual genes and measure how ‘noisy’ protein expression is and what the potential drivers of noise were, given that there are a scary amount of potential factors that can cause variation in gene expression…
The general conclusions were pretty consistent – different genes exhibit variable degrees of noise which may reflect properties of those genes: either the DNA elements that influence gene expression itself or trans factors which act on those elements
What does it mean – well if you are a bacteria it seems this noise may allow for some level of natural selection to favor the lucky few who happen to exist in a favorable state when nature calls.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1460268/
https://www.ncbi.nlm.nih.gov/pmc/artic... href=" https://pubmed.ncbi.nlm.nih.gov/15308767/ ">https://pubmed.ncbi.nlm.nih.gov/15308767/... https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2819113/">https://www.ncbi.nlm.nih.gov/pmc/artic...
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1460268/
It has a much deeper meaning which forms one of the foundations of modern synthetic biology – the ability of isogenic cells to form multicellular organisms composed of vastly heterogeneous stable cell types. After all – mammals are isogenic cells with non-genetic individuality!
The process by which these cell types arise reflects environmental signals acting on noisy gene expression to orchestrate fixed patterns of gene expression which establish diverse cellular identities.
A concrete in C elegant from @arjunrajlab shows predictable diversification of certain cell types is altered by varying levels of transcript abundance implicating transcriptional burst size/noise as a determinant of cellular differentiation. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2836165/">https://www.ncbi.nlm.nih.gov/pmc/artic...
An implication of this work is that multiple genes converge to buffer one another against environmental effects, maintaining a highly predictable pattern of cellular differentiation across different individuals.
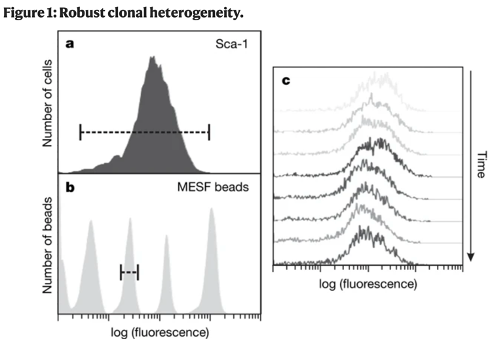
Extending this to mammals - a key study from Chang et al found lineage bias within a pool of hematopoietic progenitors could be linked to fluctuations of sca-1 protein in a ‘homogenous’ population of cells. (remember that ‘noise’ in FACS plots..)
https://www.nature.com/articles/nature06965">https://www.nature.com/articles/...
https://www.nature.com/articles/nature06965">https://www.nature.com/articles/...
By sorting low, middle, or high Sca-1 expressing cells from the population they demonstrated that each population eventually would regenerate the initial distribution (taking around 5-10 days). Suggesting this reflects semi-stable ‘noise’ in Sca-1 expression.
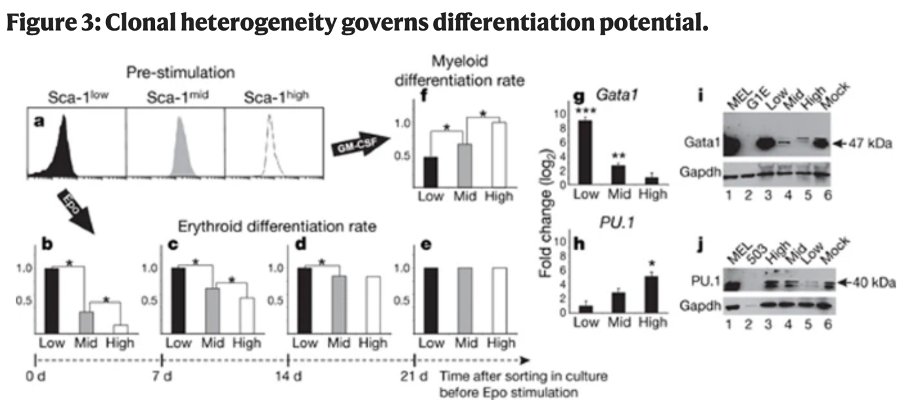
Interestingly – cells with different sca-1 expression were found to have variable capacity to generate erythroid or myeloid cells downstream of growth factors which reflects variations in GATA1 and PU.1 expression correlated to Sca-1.
Note that the timescale shifts (b-e) – so that it takes 10+ days for populations to no longer give biased differentiation despite Sca-1 showing reversion to normal distributions after 4-5 days… Variable stability across different genes?!
One of my favorite papers from Uri Alon – revisits the old temporal analysis of protein expression stability (or mixing in populations) using labeled proteins in a human cancer cell line.
Here proteins once again demonstrated patterns of heritably stable expression across multiple (2-3) cell divisions. A familiar concept emerges: ‘Such long-memory noise can act, through the cells’ regulatory circuitry, to generate variability in cellular responses and phenotypes..
These observations have now been confirmed and extended using more high throughput whole transcriptome profiling as a read out – finding hundreds of genes which show clonal stability within *isogenic populations of cancer cells. (* hard to definitively prove in cancer)
It is now increasingly appreciated that individual stem cells can exhibit highly biased patterns of differentiation in both the central nervous system and hematopoietic system.
https://www.nature.com/articles/s41593-022-01011-x">https://www.nature.com/articles/... https://www.nature.com/articles/nature12013">https://www.nature.com/articles/...
https://www.nature.com/articles/s41593-022-01011-x">https://www.nature.com/articles/... https://www.nature.com/articles/nature12013">https://www.nature.com/articles/...
In the latter there is striking evidence that this lineage bias can be heritable across cell division with clonal sisters exhibiting similar patterns of bias in different environments.
https://pubmed.ncbi.nlm.nih.gov/28235203/ ">https://pubmed.ncbi.nlm.nih.gov/28235203/... https://pubmed.ncbi.nlm.nih.gov/31974159/ ">https://pubmed.ncbi.nlm.nih.gov/31974159/...
https://pubmed.ncbi.nlm.nih.gov/28235203/ ">https://pubmed.ncbi.nlm.nih.gov/28235203/... https://pubmed.ncbi.nlm.nih.gov/31974159/ ">https://pubmed.ncbi.nlm.nih.gov/31974159/...
Does this heritability reflect short-term memory such as that described in the Sca-1 distribution? Or does it reflect heritable imprints which drive long-term non-genetic individuality within phenotypically similar cells?
Cell type diversification may be the most obvious layer of phenotypic diversity in multicellular organisms – creating a template in the form of highly regulated patterns of cellular differentiation shared by all humans
How those populations of cells are acted upon by their environment and the selective forces imposed upon them over long periods of time may be the next frontier of understanding evolution and non-genetic individuality* (sometimes referred to as ‘epigenetics’)
Identifying short-lived switches, and long-lived differences which exist within otherwise identical cells could educate all sort of future strategies to intervene in processes that are likely to be central to ageing and disease.

 Read on Twitter
Read on Twitter