New preprint. We estimate the transmissibility of #SARSCoV2 lineages based on their genomic makeup. Overall, transmissibility of #SARSCoV2 went up over time, but fitness of individual clades tends to decay due to accumulation of deleterious mutations.
1/ https://www.biorxiv.org/content/10.1101/2021.05.06.442903v1">https://www.biorxiv.org/content/1...
1/ https://www.biorxiv.org/content/10.1101/2021.05.06.442903v1">https://www.biorxiv.org/content/1...
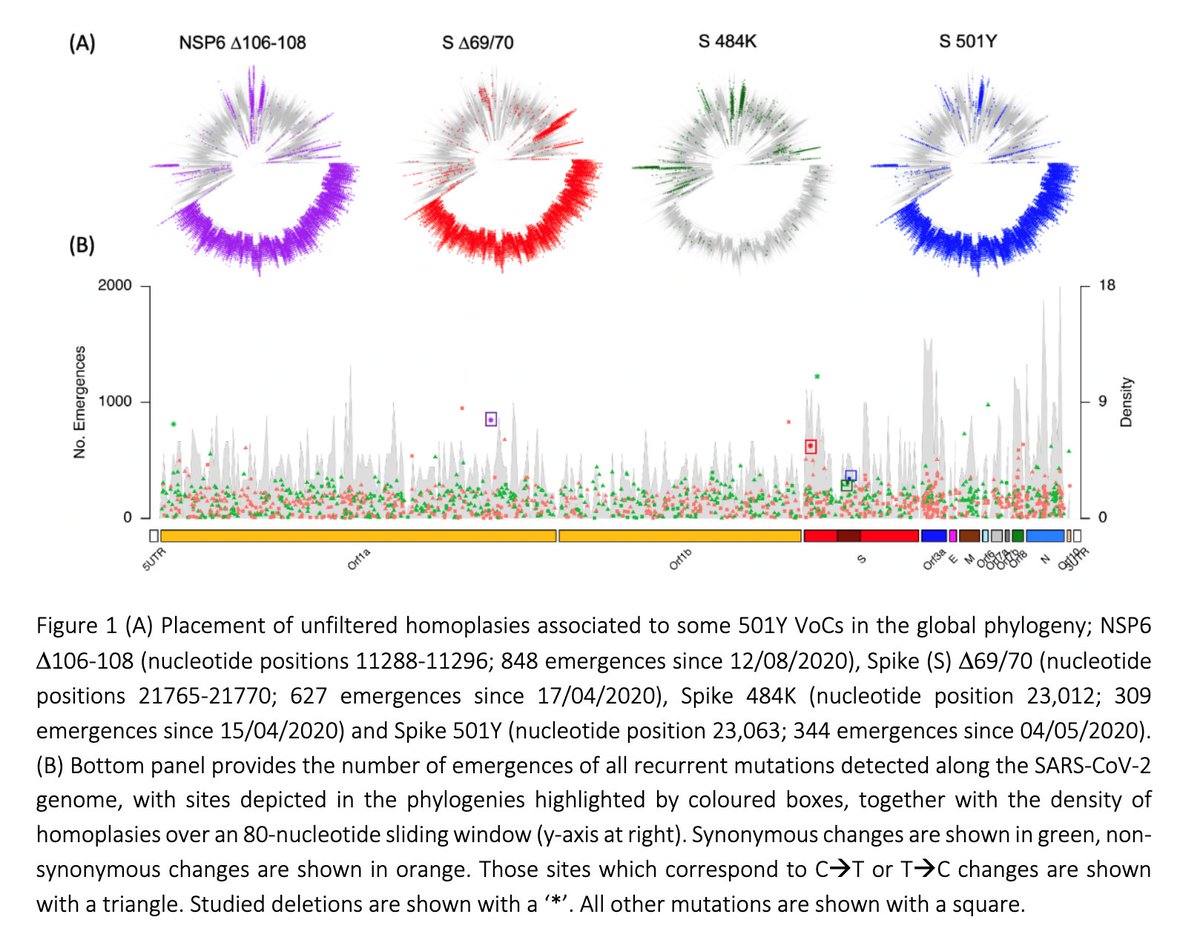
We focus on mutations / deletions that arose repeatedly as those are likely to increased viral transmission. Some arose independently hundreds of times. Below (panel A), four muts/dels associated with & #39;Variants of Concern& #39; (VoCs). B.1.1.7 clade shown 3-8 o& #39;clock in the trees.
2/
2/
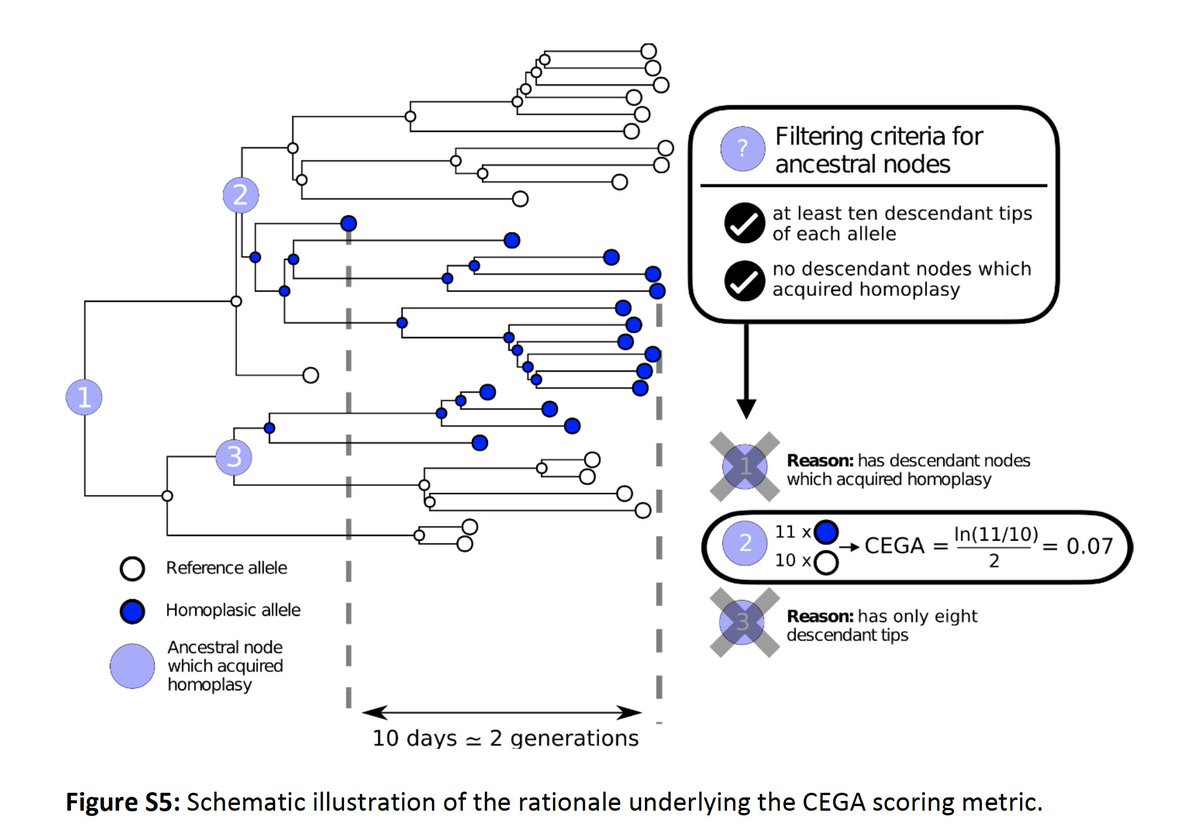
Each time a recurrent mutation appears in the #SARSCoV2 tree, we count the descendants from that node with and without the mutation and calculate the ratio of offspring of either type. We then normalise those ratios over viral generation times and average over replicates.
3/
3/
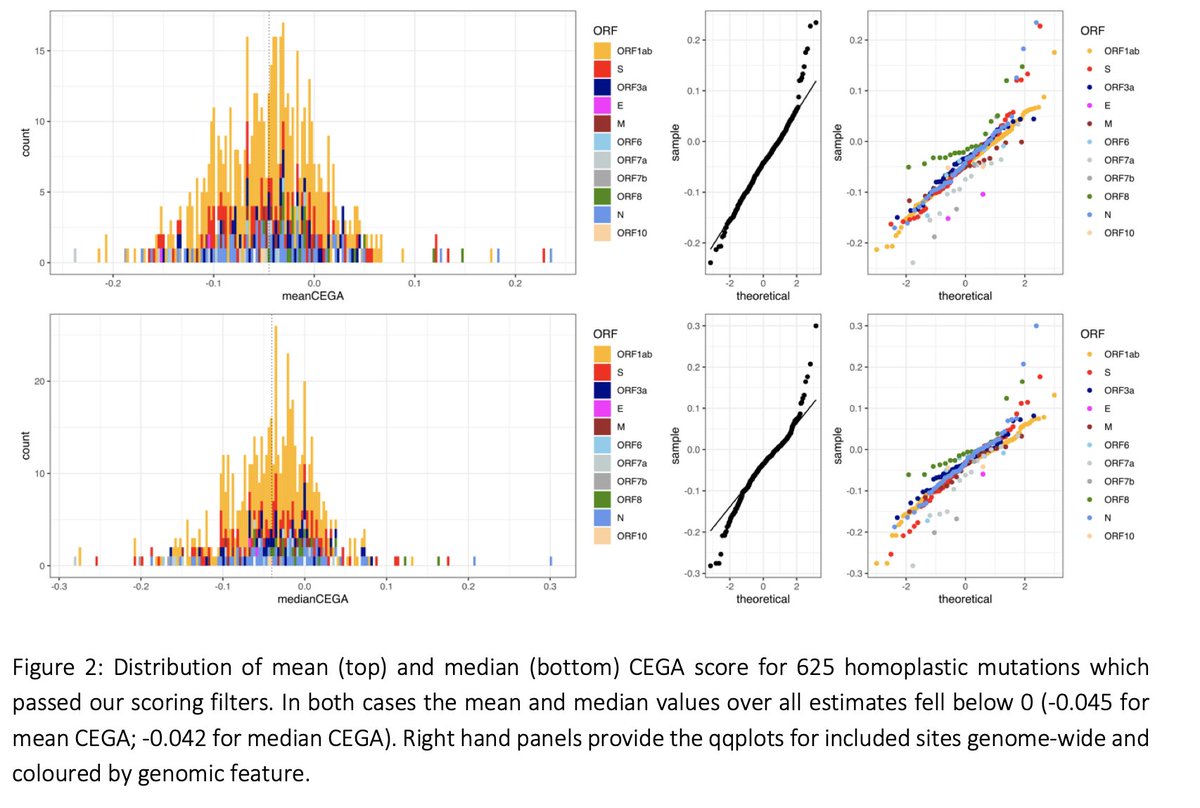
We can estimate an association with #SARSCoV2 transmissibility for 625 mutations. There is a slight tendency for mutations to be associated to reduced transmissibility overall, in particular C->T ones that are often induced by the host immune system.
4/
4/
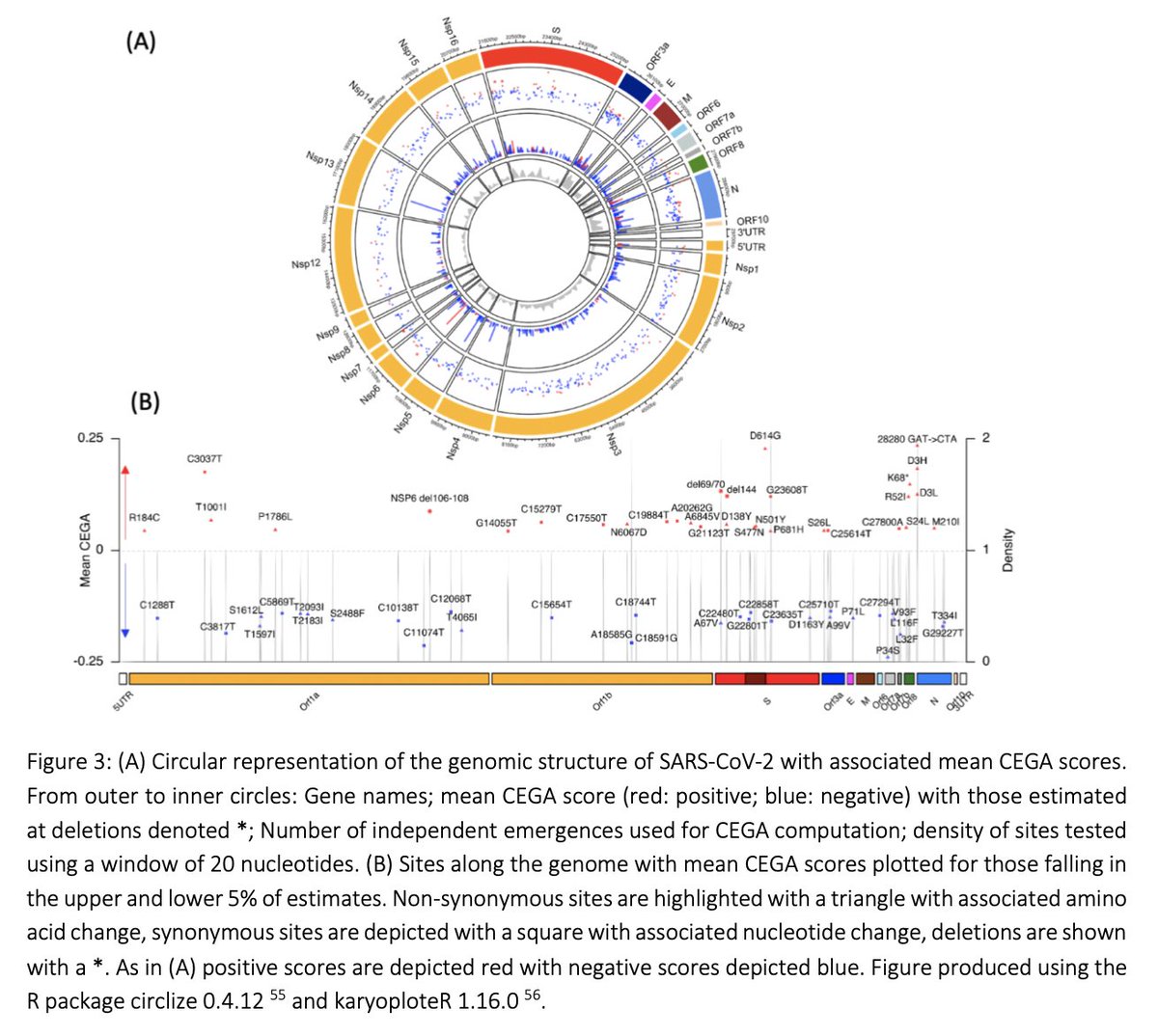
We find mutations and deletions positively associated to transmissibility throughout the #SARSCoV2 genome. Besides strong hits in the S protein, we get positive associations in NSP3, NSP6, Orf8, the nucleocapsid gene (N), and elsewhere in the genome ...
5/
5/
We next estimated the transmissibility of #SARSCoV2 strains based on the mutations / deletions they carry, under the assumption that their effect is independent of each other (i.e. disregarding possible & #39;non-linear& #39; effects of mutations).
6/
6/
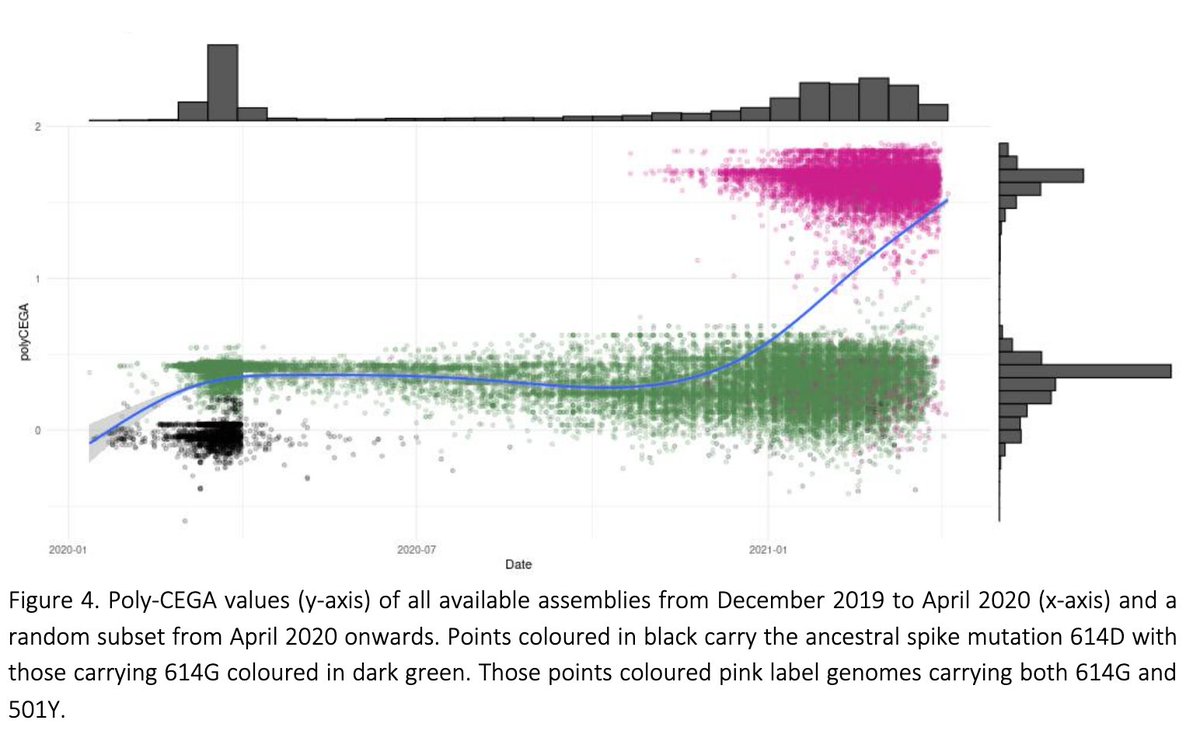
We observe two step changes in the estimated transmissibility of #SARSCoV2 over time. It first increased with the emergence of the D614G haplotype in early 2020, and then again with the emergence of the 501Y VoCs (B.1.1.7) in the second half of 2020.
7/
https://www.biorxiv.org/content/10.1101/2021.05.06.442903v1">https://www.biorxiv.org/content/1...
7/
https://www.biorxiv.org/content/10.1101/2021.05.06.442903v1">https://www.biorxiv.org/content/1...
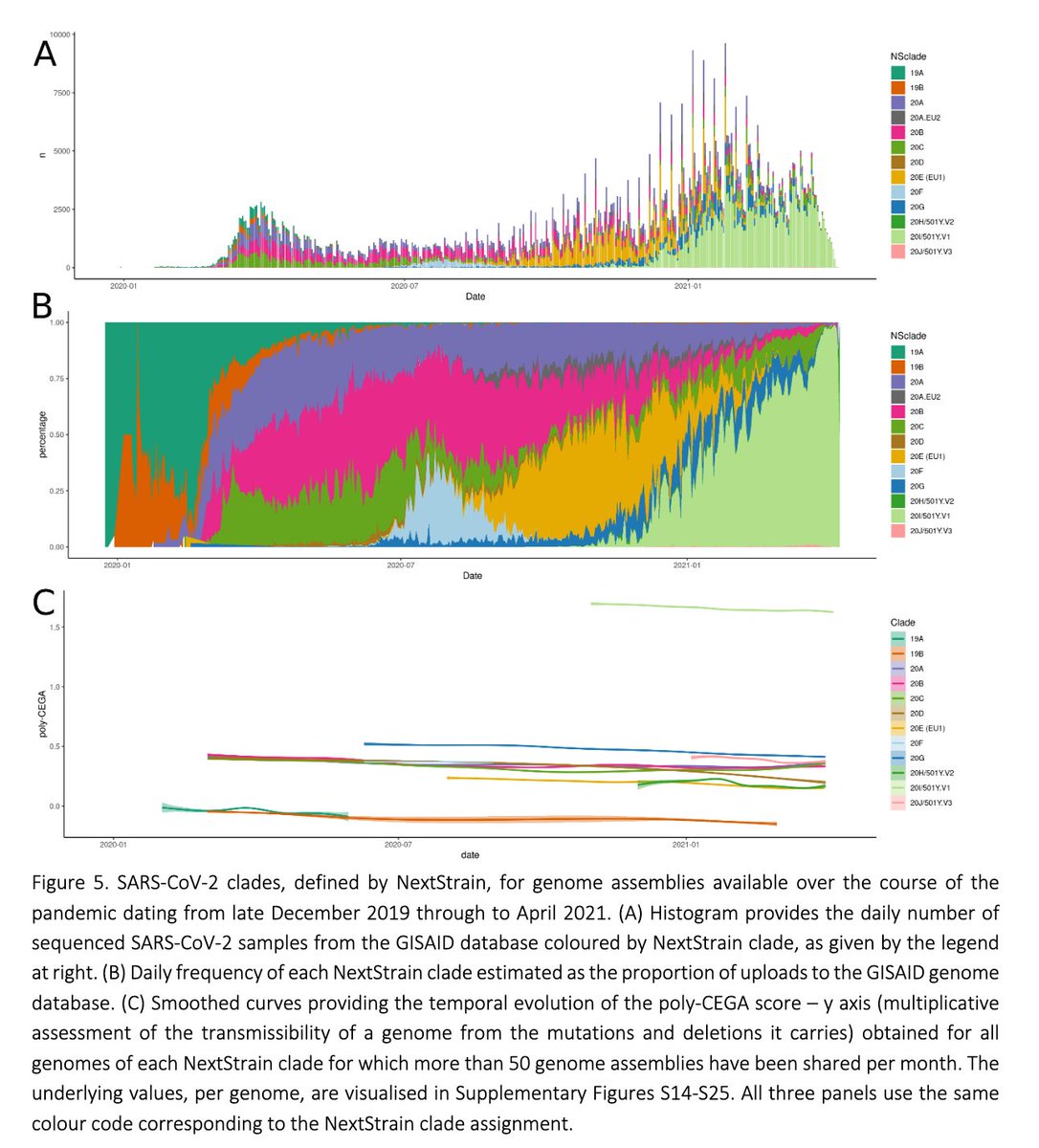
Among all major #SARSCoV2 clades, we estimate B.1.1.7 to be most transmissible. We observe a subtle, but highly consistent trend for transmissibility of #SARSCoV2 lineages to decay over time, likely due to the accumulation of deleterious mutations.
8/
https://www.biorxiv.org/content/10.1101/2021.05.06.442903v1">https://www.biorxiv.org/content/1...
8/
https://www.biorxiv.org/content/10.1101/2021.05.06.442903v1">https://www.biorxiv.org/content/1...
At this stage, our #SARCoV2 strain transmissibility estimates should be considered as relative rather than absolute. We also didn& #39;t present any result about non-linear effects (epistasis) between mutations / deletions. More work is needed for us to share those with the world.
9/
9/
To pre-empt any question about B.1.617.2, which was designated as a VoC by PHE yesterday. There is no B.1.617.2 in our dataset, but at this stage we get a fairly unremarkable preliminary transmissibility estimate for B.1.617 of ~0.33.
10/
10/
This analysis was enabled by the GISAID EpiCoV database ( https://www.gisaid.org"> https://www.gisaid.org ). We gratefully acknowledge all contributing / submitting labs around the globe including COG-UK ( https://www.cogconsortium.uk"> https://www.cogconsortium.uk ) who have openly shared large numbers of UK SARS-CoV-2 assemblies.
11/
11/
Finally, I want to thank everyone in our wonderful team, Damien Richard, Liam Shaw, @CedricCSTan1, @ChrisJ_Owen, @misac42, our guest star @RobLanfear and my wonderful colleague @LucyvanDorp.
12/ https://www.biorxiv.org/content/10.1101/2021.05.06.442903v1">https://www.biorxiv.org/content/1...
12/ https://www.biorxiv.org/content/10.1101/2021.05.06.442903v1">https://www.biorxiv.org/content/1...

 Read on Twitter
Read on Twitter