Excited to share our latest work. We present NanoSeq, a new duplex sequencing method with <5 errors per billion sites in single DNA molecules. A leap in our ability to study mutation in any tissue. 1/ https://www.nature.com/articles/s41586-021-03477-4">https://www.nature.com/articles/...
Somatic mutations are important to study cancer and ageing, but detecting them in normal tissues has been very challenging as they are often only present in single cells or small groups of cells (clones). 2/
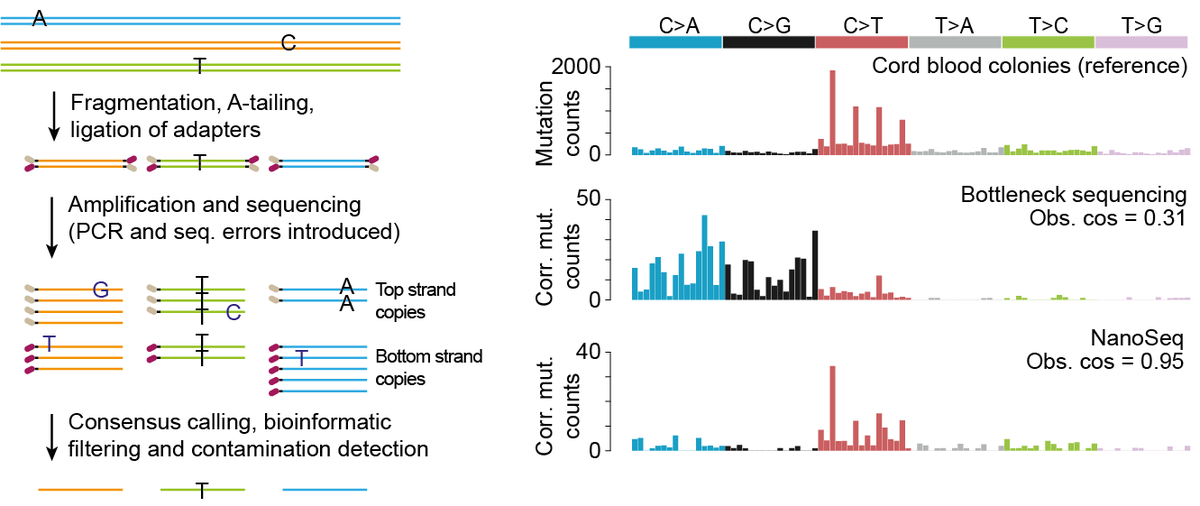
Duplex sequencing (2012) and bottleneck sequencing (2016) are amazing technologies, achieving error rates <1 error per million sites in single DNA fragments, thanks to sequencing both strands of a DNA molecule independently. 3/ https://www.pnas.org/content/109/36/14508">https://www.pnas.org/content/1...
Over 4 years of iterative refinement, Rob Osborne and @AbascalFed identified sources of error in duplex sequencing: end repair, nick extension, mapping errors, DNA contamination… devising solutions to each of them and eventually achieving rates <5 errors per billion sites. 4/
This error rate is two orders of magnitude lower than the typical mutation rates of human tissues, enabling the study of somatic mutagenesis in any tissue or population of cells, independently of clonality, from as little as 1ng of DNA. 5/
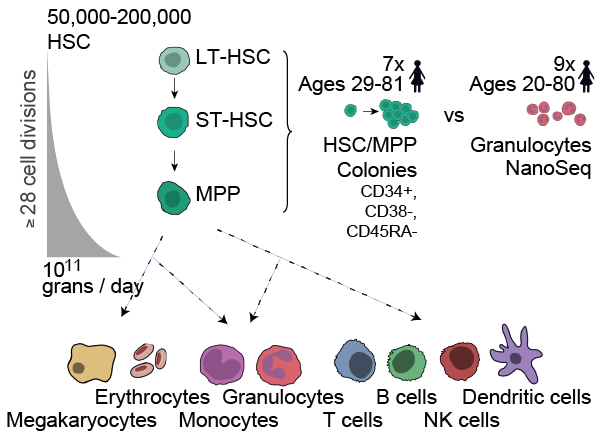
For the first time, we could reliably study somatic mutagenesis in any cell type, even in non-dividing cells. We decided to address 2 questions: (1) compare differentiated cells and stem cells in mitotic tissues, (2) sequence post-mitotic tissues. 6/
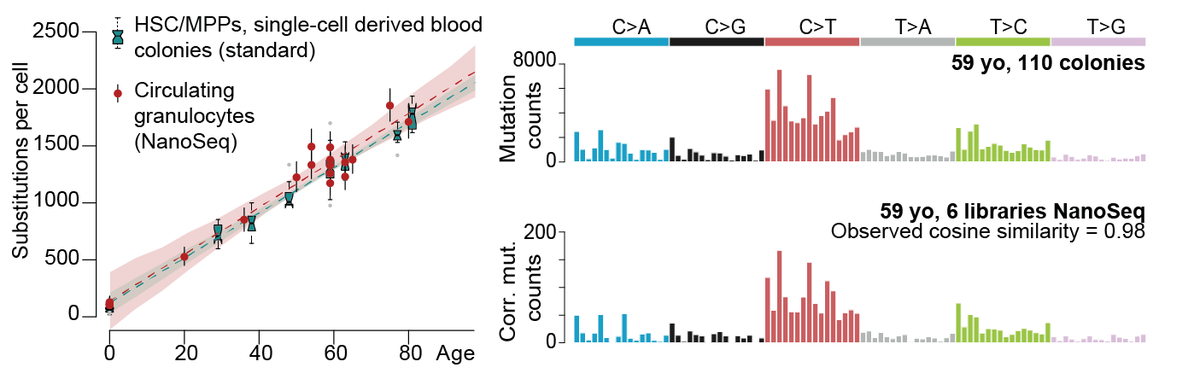
Haematopoietic stem cells (HSCs) divide rarely (~once per year), but sustain an enormous production of blood cells thanks to the high proliferation of intermediate progenitors. Given estimates of division rates, we expected many more mutations in granulocytes than HSCs. 7/
NanoSeq revealed that a considerable number of additional cell divisions does not seem to translate into a comparably higher number of mutations. We then sequenced post-mitotic neurons and rarely dividing cells from smooth muscle. 8/
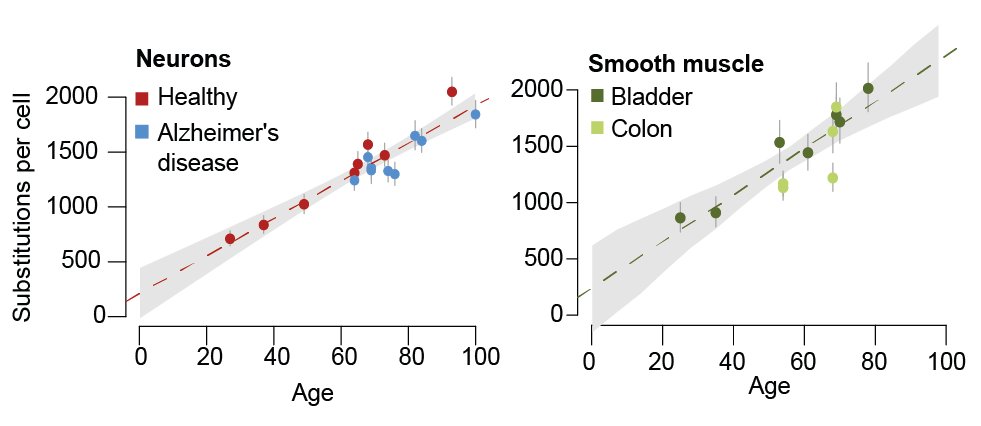
We found that neurons accumulate mutations linearly throughout life in the absence of cell division, in line with previous single-cell sequencing data. Smooth muscle too, which had remained unstudied. 9/
Overall, we found similar mutation rates and signatures in post-mitotic cells and largely non-dividing cells than in some mitotic tissues, arguing against cell division as the dominant source of somatic mutations (at least for SBS5 and SBS16). 10/
The ability to reliably detect mutations in single DNA molecules has much wider applications. It could greatly facilitate the study of mutagenesis in vitro and in vivo, screening for new carcinogens and performing somatic mutation studies in large cohorts non-invasively. 11/
This is the creation of the amazing Rob Osborne and @AbascalFed. But this is also truly a team effort, with key contributions from @_Luke_MRH_, Emily Mitchell and @andrewrjlawson on the biology, as well as Stef Lensing and Pete Ellis from @sangerinstitute R&D. 12/
And from other brilliant colleagues, including @R_Rahbari, @KSaebParsy, @AndyRusss, Peter Campbell, Mike Stratton and all other authors, which supported this work in many ways. Thank you also to the donors and their families and to @MT_Genetics, @wellcometrust and @CRUKresearch .
Free access to the paper here: https://rdcu.be/cjBtH .">https://rdcu.be/cjBtH&quo... The detailed protocol can be accessed here: http://dx.doi.org/10.21203/rs.3.pex-1298/v1.">https://dx.doi.org/10.21203/... And all of the bioinformatic code here: http://dx.doi.org/10.5281/zenodo.4604537.">https://dx.doi.org/10.5281/z...

 Read on Twitter
Read on Twitter