I do like that $GNCA uses autologous dendritic cells to expand their blood derived T cells. It& #39;s easy to see patient specific mutations but hard to ensure that mutations are true neoantigens, i.e. to be able to be presented by MHC and recognized by TCRs. DC& #39;s are insurance.
The three basic ways of ensuring your TIL-like product targets recognizable neoantigens:
1) Let the specific tumor select the T cells. $IOVA
2) Use external non-patient specific info and algos. $ACHL
3) Use patient specific dendritic cells to selectively expand T cells. $GNCA
1) Let the specific tumor select the T cells. $IOVA
2) Use external non-patient specific info and algos. $ACHL
3) Use patient specific dendritic cells to selectively expand T cells. $GNCA
Again, the problem is that the majority of mutated proteins are either not presentable by MHC, or if presentable, not recognized by T cells.
I prefer methods that use a patient& #39;s own flesh and cells to ensure that mutations are true neoantigens.
I prefer methods that use a patient& #39;s own flesh and cells to ensure that mutations are true neoantigens.
I& #39;m most skeptical about $ACHL process. They pick neoantigens from two info sources: a) patient specific sequencing b) database of neoantigens from patients w/ same tumor type. Look for overlap.
There is no patient specific guarantee that these neoantigens are recognizable.
There is no patient specific guarantee that these neoantigens are recognizable.
Lets assume you& #39;ve infused neoantigen specific T cells. How important is it that that neoantigens are expressed on all tumor cells? $ACHL think v important. I think nice, but luxury. More important to maximize probability your T cells will target and kill at least some cells.
I like $IOVA, like TILS, do NOT like next-gen approaches that sacrifice the #1 advantage of old-school TILS, which is:
they maximize the likelihood that subset of infused TILs will target at least subset of tumor cells. Max chance for some response, antigen spreading, etc.
they maximize the likelihood that subset of infused TILs will target at least subset of tumor cells. Max chance for some response, antigen spreading, etc.
For anything next-gen TILs or TIL-like approaches I think that& #39;s the #1 Q: is this approach reducing the likelihood that at least a subset of T cells will target at least a subset of tumor cells?
Don& #39;t forego info provided by real activation of autologous T cells by APCs/tumor.
Don& #39;t forego info provided by real activation of autologous T cells by APCs/tumor.
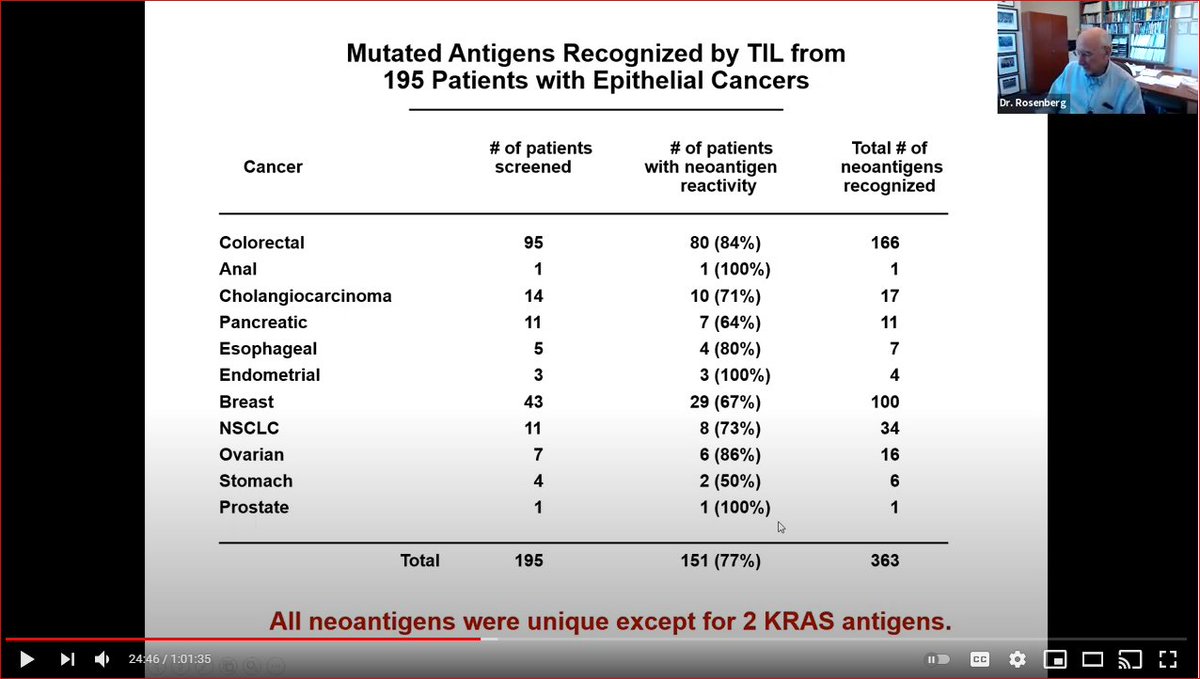
What neoantigens do TILs recognize when TILs work?
Rosenberg
-random: few shared across patients
-random 2: mostly NOT "driver" mutations
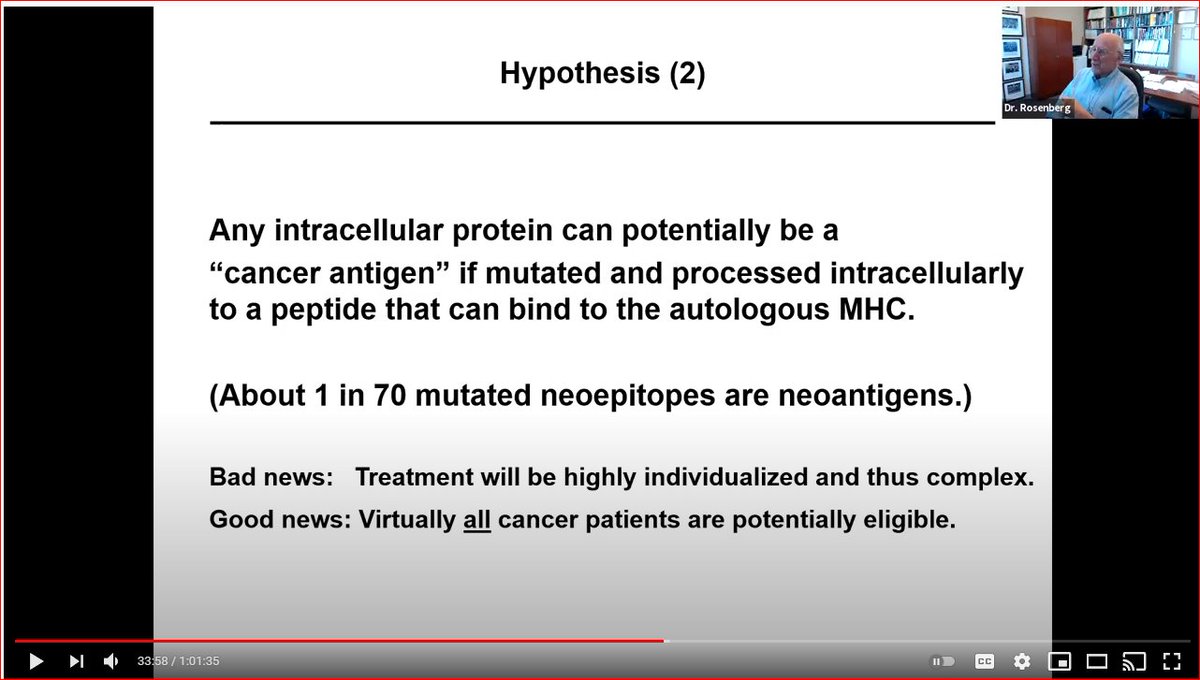
-rare, i.e. 1/70 neoepitopes are neoantigens
-but all patients have true neoantigens
Implication? Leverage all patient specific info.
Rosenberg
-random: few shared across patients
-random 2: mostly NOT "driver" mutations
-rare, i.e. 1/70 neoepitopes are neoantigens
-but all patients have true neoantigens
Implication? Leverage all patient specific info.
Again, the implication of neoantigens being super random and hard to predict from theory is that you must leverage all patient specific info. Not just tumor sequencing. But also info from actual T/APC/tumor interactions. https://www.youtube.com/watch?v=IbPBCFrGs_M">https://www.youtube.com/watch...

 Read on Twitter
Read on Twitter