If any of your professors/advisors ever give you this advice, please remember that many students are dependent on tools/techniques which were readily available in the 1980s.
We can use @Stylopidae& #39;s dissertation as an example. Out of 6 chapters, only 2 depend on newer tech. https://twitter.com/nanophononics/status/1384672634340712462">https://twitter.com/nanophono...
We can use @Stylopidae& #39;s dissertation as an example. Out of 6 chapters, only 2 depend on newer tech. https://twitter.com/nanophononics/status/1384672634340712462">https://twitter.com/nanophono...
Chapter 1: We grew sugarbeets in buckets with grass rings around them, and trimmed grass to ensure the beets weren& #39;t shaded. We then removed the grass at specific timepoints
The newest technology involved in this study were plastic bags, invented in 1950
https://www.plasticsmakeitpossible.com/about-plastics/history-of-plastics/plastic-innovations-in-packaging-through-the-decades/">https://www.plasticsmakeitpossible.com/about-pla...
The newest technology involved in this study were plastic bags, invented in 1950
https://www.plasticsmakeitpossible.com/about-plastics/history-of-plastics/plastic-innovations-in-packaging-through-the-decades/">https://www.plasticsmakeitpossible.com/about-pla...
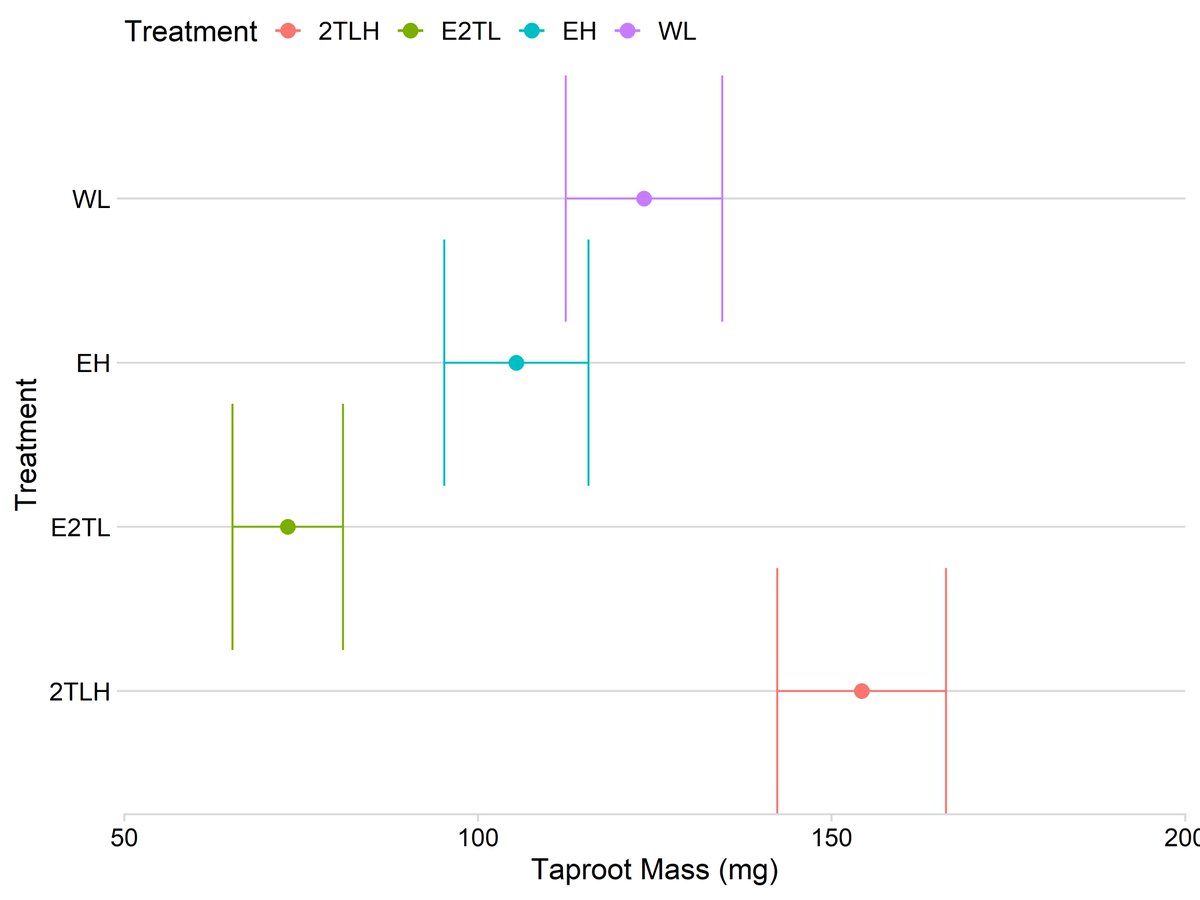
Chapter 2: We wanted to see if water would change the ability of sugarbeets to get bigger while growing close to other plants.
So instead of adding/removing grass, as we did in chapter 1, we simply poked holes in the trash bag.
Newest technology: 1950
So instead of adding/removing grass, as we did in chapter 1, we simply poked holes in the trash bag.
Newest technology: 1950
Chapter 3: We want to see if sugarbeets have trouble getting bigger when we grow them too close to other plants IRL.
So instead of shielding them with trash bags, we used aluminum siding.
Aluminum is as good as copper, so the newest technology here is from 5000 BCE
So instead of shielding them with trash bags, we used aluminum siding.
Aluminum is as good as copper, so the newest technology here is from 5000 BCE
The next few chapters are looking at the size and shape of leaves and cotyledons of sugarbeets grown under full-spectrum (WL), Far-red (FR) and dark conditions...and switching between the two.
Nowadays, this is done with ImageJ but...
Nowadays, this is done with ImageJ but...
...the precursor to ImageJ was a program called ObjectPascal developed in the early 1980s and commercialized in 1986.
The majority of this dataset, however, could be inferred from data you can get from a simple ruler.
So newest technology: 1986...just before @Stylopidae
The majority of this dataset, however, could be inferred from data you can get from a simple ruler.
So newest technology: 1986...just before @Stylopidae
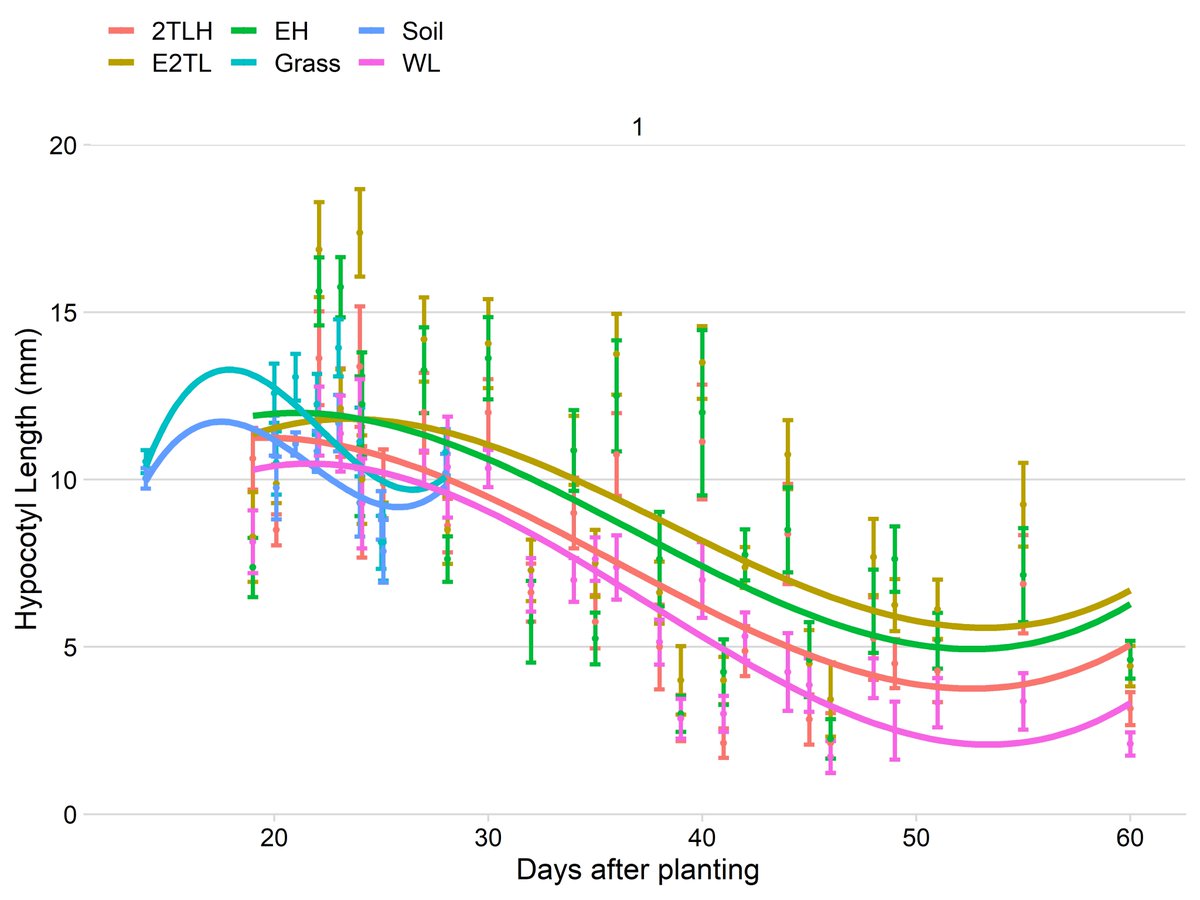
The most interesting finding we& #39;ve had, however, is about the growth of the area between the soil and the first two leaf-like things you see when a plant emerges (cotyledons).
We& #39;re not sure if that hypocotyl tissue incorporates itself into the root...
We& #39;re not sure if that hypocotyl tissue incorporates itself into the root...
...but, again, this is all done using a ruler.
Newest technology: The metric system, invented in 1790 (with a nod to the use of computers in the late 1980s, which fulfills the OP& #39;s challenge).
Newest technology: The metric system, invented in 1790 (with a nod to the use of computers in the late 1980s, which fulfills the OP& #39;s challenge).
It& #39;s also worth noting that there WERE analysis of leaf shapes which took place in the 1980s.
These were not common, however, with the proper collaborations (usually the maths folks), they were more than possible.
https://cdnsciencepub.com/doi/abs/10.1139/b83-256">https://cdnsciencepub.com/doi/abs/1...
These were not common, however, with the proper collaborations (usually the maths folks), they were more than possible.
https://cdnsciencepub.com/doi/abs/10.1139/b83-256">https://cdnsciencepub.com/doi/abs/1...
Nowadays, however, this same stuff is taken care of using automated leaf-measuring software.
This is using LeafJ, developed by Julian Maloof in 2013.
So, yes, things have gotten more efficient...but everything was already there in the 1980s.
This is using LeafJ, developed by Julian Maloof in 2013.
So, yes, things have gotten more efficient...but everything was already there in the 1980s.
I would argue that all of this research could have progressed in a similar timeframe in 1987, but the paper would probably have a few more authors...and my author order might have changed.
Either way, definitely possible.
Either way, definitely possible.
Chapter 6 is the only one for which I do not have preliminary data. This is a microscopy experiment, building on a paper published in 2020.
Jammer, A. et. al (2020). Early‐stage sugar beet taproot development is characterized by three distinct physiological phases. Plant direct
Jammer, A. et. al (2020). Early‐stage sugar beet taproot development is characterized by three distinct physiological phases. Plant direct
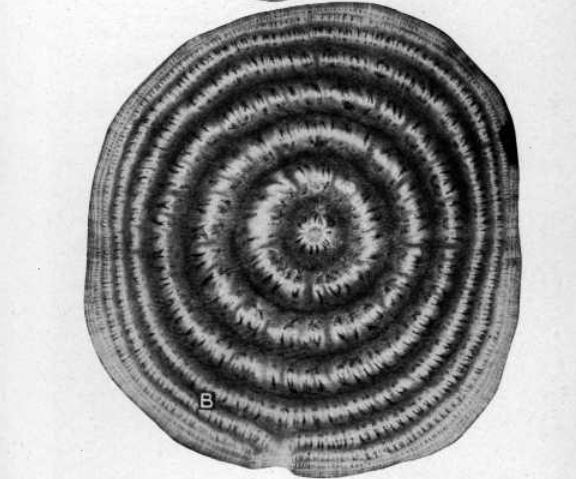
The stain used to generate the picture above is a mix called "Etzholdt& #39;s solution", which was first published in 2002.
The protocol this team was building on, however, was developed in 1926.
The stain just makes it easier to tell xylem and phloem apart.
The protocol this team was building on, however, was developed in 1926.
The stain just makes it easier to tell xylem and phloem apart.
This is an image generated by Ernst Artschwager in 1926. The details would take an entire thread, but I can differentiate between these tissues just by looking at this picture.
Yes, it& #39;s a bit more difficult, but the photos in the 1930s weren& #39;t meant for Twitter.
Yes, it& #39;s a bit more difficult, but the photos in the 1930s weren& #39;t meant for Twitter.
Earliest technology: :1926.
I& #39;m being VERY generous here, because we& #39;ve been able to obtain these images (via hand-tracing, mind you), since 1644
Artschwager, E. (1926). Anatomy of the vegetative organs of the sugar beet. Journal of Agricultural Research, 33, 143–176.
I& #39;m being VERY generous here, because we& #39;ve been able to obtain these images (via hand-tracing, mind you), since 1644
Artschwager, E. (1926). Anatomy of the vegetative organs of the sugar beet. Journal of Agricultural Research, 33, 143–176.
The ONE chapter from @Stylopidae& #39;s dissertation which wasn& #39;t possible to do in the 1980s is the 7th, and that is not actually meant for publication.
It& #39;s a grant proposal.
We& #39;re still going to get publishable data from this, but it& #39;s not actually meant to be published.
It& #39;s a grant proposal.
We& #39;re still going to get publishable data from this, but it& #39;s not actually meant to be published.
Originally, this was lab-lore. There was a vague idea that plants grown too close to one another were unsuitable for science.
@IdahoWeeds started the first and second chapters by asking whether this was an actual thing, and whether this could be an economic problem.
@IdahoWeeds started the first and second chapters by asking whether this was an actual thing, and whether this could be an economic problem.
Which, BTW, follow Albert and ask all your planty questions. Also, @WyoWeeds.
These are both fantastic scientists, and great mentors.
ANYWAYS...
These are both fantastic scientists, and great mentors.
ANYWAYS...
We haven& #39;t gotten to chapter 7 yet, behind the scenes, but we& #39;re hoping to start SOMETHING on this next week.
But here& #39;s where I& #39;m going to be REALLY generous
Microarray technologies, used by folks like @23andMe , were only developed in the LATE 1990s. Usable only in early 2000
But here& #39;s where I& #39;m going to be REALLY generous
Microarray technologies, used by folks like @23andMe , were only developed in the LATE 1990s. Usable only in early 2000
@23andMe, when it comes to health, to be VERY frank, is kind of bullshit.
A lot of people view 23andMe as their only resort for genetic sequencing, but they use microarrays...and this is not a valid testing method. https://www.nature.com/news/the-rise-and-fall-and-rise-again-of-23andme-1.22801">https://www.nature.com/news/the-...
A lot of people view 23andMe as their only resort for genetic sequencing, but they use microarrays...and this is not a valid testing method. https://www.nature.com/news/the-rise-and-fall-and-rise-again-of-23andme-1.22801">https://www.nature.com/news/the-...
If there& #39;s enough interest, we& #39;d be happy to do a #DeepDive.
Basically, they use a technology that depends on the ability of gene fragments to bind to other gene fragments on a microchip.
DEFINITELY useable for some kinds of parental testing/ID, sequencing is needed for health.
Basically, they use a technology that depends on the ability of gene fragments to bind to other gene fragments on a microchip.
DEFINITELY useable for some kinds of parental testing/ID, sequencing is needed for health.
To get this grant, we need to do another set of studies (Chapters 4 and 5 basically) on a genetic model plant.
I& #39;d argue the technologies needed to GET these funds were around in the late 1980s, but the techniques we want to use were not around before 2000.
I& #39;d argue the technologies needed to GET these funds were around in the late 1980s, but the techniques we want to use were not around before 2000.
So, here& #39;s a modern PhD dissertation (publication anticipated by 2023) which relies entirely on techniques which were available before 1989.
While modern technology HAS made this stuff easier, before 2020, it was always possible to take pictures and hand it off to collaborators.
While modern technology HAS made this stuff easier, before 2020, it was always possible to take pictures and hand it off to collaborators.
In many ways, plants are MUCH easier than insects...because plants have characteristics easier captured by photography.
Perennial Friends of the Blog @BombusChristy and @RvingNaturalist rely a lot more on tools which are far older.
Colored bowls and insect species keys.
Perennial Friends of the Blog @BombusChristy and @RvingNaturalist rely a lot more on tools which are far older.
Colored bowls and insect species keys.
The fact of the matter is that modern PhD candidates are expected to publish more, in a harder publishing environment, to get a chance in succeeding in a far more competitive job market than the OP ever faced.
The most intelligent-and capable-scientists who have ever lived, are the PhD students doing research RIGHT NOW. In the modern age.
Yes, we are building on the people who came before us...but the standards are almost unachievably high.
Yes, we are building on the people who came before us...but the standards are almost unachievably high.
@Stylopidae& #39;s dad did his PhD in the early 1980s, and it was a respectable dissertation back then, looking at drought tolerance in corn.
It yielded ZERO publications, yet, it& #39;s still cited today.
It yielded ZERO publications, yet, it& #39;s still cited today.
In the modern era, it would be comparable to a MSc dissertation in the early/late 2010s.
A PhD dissertation with zero publications, today, would be indefensible. It simply wouldn& #39;t pass, and would be considered a hallmark of a bad student/incompetent mentor.
A PhD dissertation with zero publications, today, would be indefensible. It simply wouldn& #39;t pass, and would be considered a hallmark of a bad student/incompetent mentor.
So, if an advisor, who got their job off 2-3 publications, tells you that your job is harder than theirs...well, that simply isn& #39;t true.
The standards are simply much, much, much higher for modern scientists.
The standards are simply much, much, much higher for modern scientists.

 Read on Twitter
Read on Twitter