How can we systematically measure gene regulatory networks around key disease genes?
Excited to share our new work using CRISPR KOs in primary T cells, with @MarsonLab, led by @JakeFreimer and Oren Shaked: https://www.biorxiv.org/content/10.1101/2021.04.18.440363v1">https://www.biorxiv.org/content/1...
Excited to share our new work using CRISPR KOs in primary T cells, with @MarsonLab, led by @JakeFreimer and Oren Shaked: https://www.biorxiv.org/content/10.1101/2021.04.18.440363v1">https://www.biorxiv.org/content/1...
There are now many 1000s of GWAS hits, but connecting hits to function remains difficult. There are various approaches to connect SNPs to causal genes in cis -- but it& #39;s still much harder to determine how the cis-genes may affect downstream targets via trans-regulation.
There are still only a few examples where it has been possible to determine downstream pathways from GWAS hits: eg IRX3/5 from Melina Claussnitzer/Marcelo Nobrega; KLF14 from Mark McCarthy. But new perturbation approaches in primary cells present a promising way forward.
Jake and Oren developed a systematic approach to (1) identify upstream regulators of known autoimmune genes using CRISPR screening, and (2) identify downstream members of the same gene networks by CRISPR KO+ RNA-seq
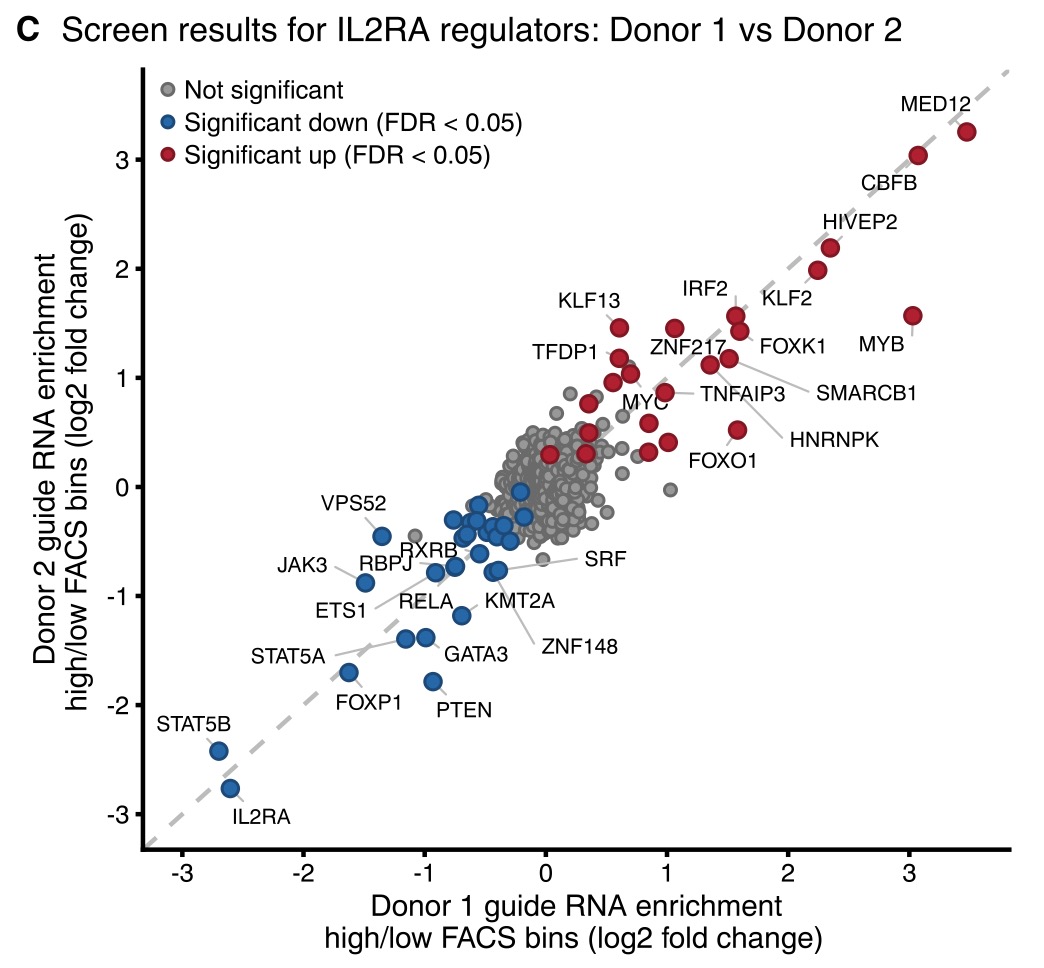
Starting from IL2RA, IL-2, and CTLA4, they performed CRISPR-screening to identify upstream regulators in primary CD4+ T cells. Results were extremely clean and reproducible -- shown here comparing 2 donors for IL2RA:
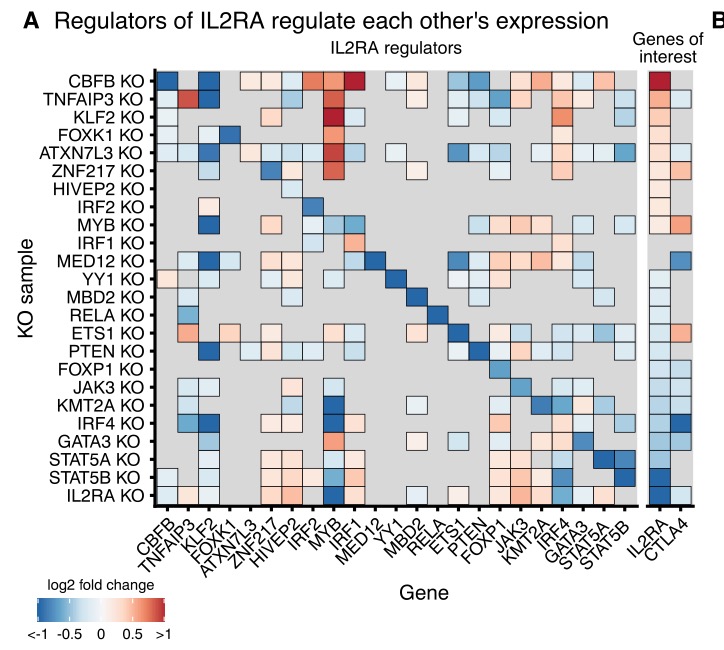
They then selected 24 upstream regulators for arrayed KO followed by RNA-seq and ATAC-seq. This works starts to reveal a highly interconnected network among the upstream regulators and downstream targets.
Here& #39;s the matrix of significant effects among the regulators:
Here& #39;s the matrix of significant effects among the regulators:
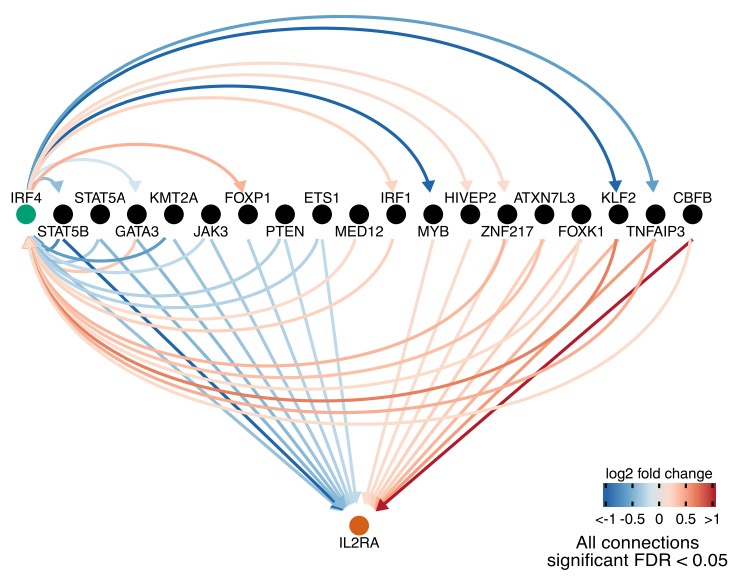
One striking observation is the high prevalence of feedback loops within the set of regulators: for example between IRF4 and other regulators of IL2RA
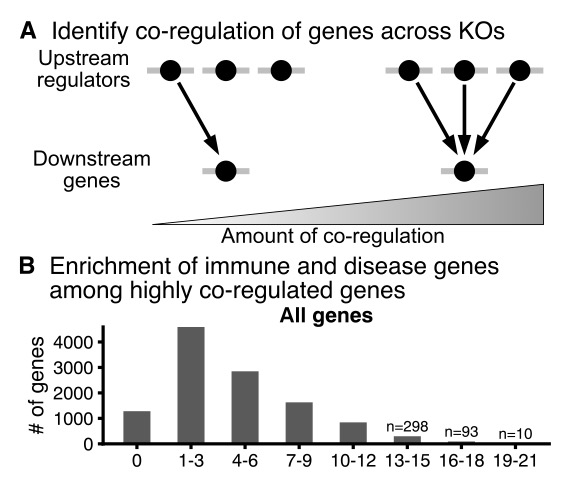
Next, Jake hypothesized that other *downstream* genes that are affected by KO of IL2RA regulators might also be enriched for immune functions. For every expressed gene we can ask how many KOs affect its expression (0 to max possible 24):
Turns out that genes that are most highly plugged into this IL2RA network are *hugely* enriched for immune functions. To be honest I was kind of blown away by how well this worked.
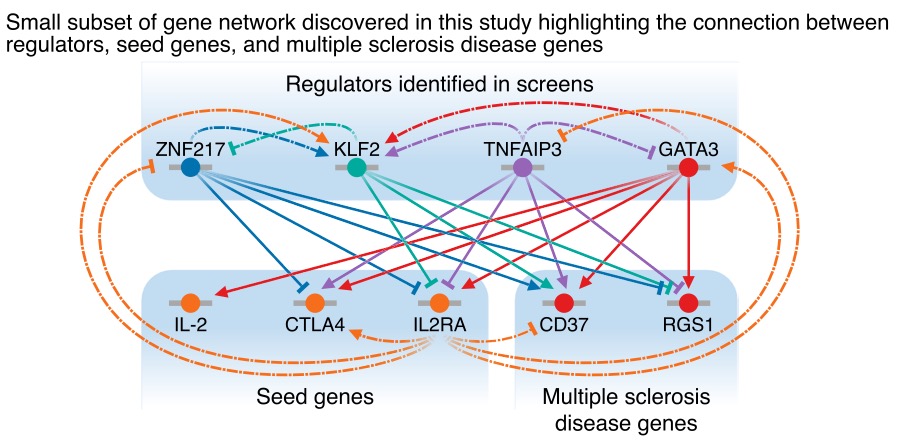
We can also start to map out networks around specific diseases and disease genes. Eg multiple sclerosis hits were particularly enriched in the IL2RA network. Here is a part of that network (GATA3, CD37 and RGS1 all have MS hits):
In summary, it& #39;s becoming clear that perturbation-based approaches can be an important tool for dissecting trans-regulatory networks for complex traits. Thanks to work by @MarsonLab and other labs in the field we can expect huge progress in this space.
An analogy is that finding cis effects of variants on genes is like trying to understand commuting patterns in NY by determining everyone& #39;s nearest subway station: helpful, but v limited.
KO allows us to close stations and measure effects on traffic through every other station.
KO allows us to close stations and measure effects on traffic through every other station.
Most important, this was a wonderful collaboration with @MarsonLab and I can& #39;t say enough about the amazing, creative work by @JakeFreimer and Oren Shaked to make this project happen.

 Read on Twitter
Read on Twitter