Yes, a neuroscientist can understand a microprocessor...a rebuttal thread on the popular paper by @stochastician and @KordingLab
While I agree with some of the points, I worry the paper leaves a misleading impression about neuroscience experiments 1/23 https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1005268">https://journals.plos.org/ploscompb...
While I agree with some of the points, I worry the paper leaves a misleading impression about neuroscience experiments 1/23 https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1005268">https://journals.plos.org/ploscompb...
I agree that just creating more complex datasets and analytics is unlikely to result in insights we need.
But in making the argument it:
1. casually dismisses large body of existing knowledge
2. doesn& #39;t recognize this was obtained by skillfully probing the brain 2/23
But in making the argument it:
1. casually dismisses large body of existing knowledge
2. doesn& #39;t recognize this was obtained by skillfully probing the brain 2/23
"used naively" is the problem.
A large set of neuroscience experiments exist that:
1.are hypothesis driven
2.control for confounds
3.apply techniques correctly, not naively.
Resulting in observations one can be confident about, even if the conclusions might change 3/
A large set of neuroscience experiments exist that:
1.are hypothesis driven
2.control for confounds
3.apply techniques correctly, not naively.
Resulting in observations one can be confident about, even if the conclusions might change 3/
Applying even the rudimentary techniques correctly would have resulted in a far greater understanding of the microprocessor than what is implied.
Many experimental neuroscientists are not that different from electrical engineers in how they approach the problem.
Let& #39;s dig in 4/
Many experimental neuroscientists are not that different from electrical engineers in how they approach the problem.
Let& #39;s dig in 4/
Paper analyzed
1.the connections on the chip,
2.the effects of destroying individual transistors,
3.single-unit tuning curves,
4.the joint statistics across transistors,
5.local activities, ..and more.
Let& #39;s look at at some of these.. 5/
1.the connections on the chip,
2.the effects of destroying individual transistors,
3.single-unit tuning curves,
4.the joint statistics across transistors,
5.local activities, ..and more.
Let& #39;s look at at some of these.. 5/
Connections:
Here the paper goes full connectomics and big data. But that is not how a lot of neuroanatomy work is done. Many Neuroscientists explicitly probe for connectivity between certain elements because they have hypotheses driving those experiments. 6/
Here the paper goes full connectomics and big data. But that is not how a lot of neuroanatomy work is done. Many Neuroscientists explicitly probe for connectivity between certain elements because they have hypotheses driving those experiments. 6/
Take works from @mattlark, Angelucci, Thompson, etc. resulting in a wealth of information (Note I don& #39;t call this wealth of data) about connectivity& physiology, with good qns and good techniques.
Here is an example of a paper describing such work: https://academic.oup.com/cercor/article/13/1/5/354630">https://academic.oup.com/cercor/ar...
Here is an example of a paper describing such work: https://academic.oup.com/cercor/article/13/1/5/354630">https://academic.oup.com/cercor/ar...
Another example is cytochrome oxidase blobs. It started with the observation that a set of neurons in v1 stain differently, and scientists have pulled on this thread in very ingenious ways to find organizational properties of neocortical circuits https://pubmed.ncbi.nlm.nih.gov/28077720/ ">https://pubmed.ncbi.nlm.nih.gov/28077720/...
It is easy to dismiss these as mere stamp collecting, (Not a word used in the paper, but often used to dismiss experimental work) but I think such experiments do provide clues regarding computational organization.
I think the paper missed on a valid criticism of connectomics: It will be large scale, but overall less reliable than the findings above as connectome won& #39;t be 100% accurate.
OTOH, the paper is right that many connectome findings are likely to be of a statistical nature.
OTOH, the paper is right that many connectome findings are likely to be of a statistical nature.
Lesion studies:
IMO lesion studies in the paper were a mischaracterization. I haven& #39;t seen many neuroscience papers just randomly lesioning neurons. Often the lesion studies test for specific hypotheses based on the existing knowledge, in controlled experimental settings.
IMO lesion studies in the paper were a mischaracterization. I haven& #39;t seen many neuroscience papers just randomly lesioning neurons. Often the lesion studies test for specific hypotheses based on the existing knowledge, in controlled experimental settings.
Single unit tuning curves:
Not being able to obtain informative tuning curves from a microprocessor doesn& #39;t invalidate tuning curves in the brain, provided they are interpreted appropriately. Although the paper hedges, it still strongly insinuates.
Not being able to obtain informative tuning curves from a microprocessor doesn& #39;t invalidate tuning curves in the brain, provided they are interpreted appropriately. Although the paper hedges, it still strongly insinuates.
For eg, for retinotopically organized cortical regions, neurons are dedicated to processing parts of space. Color-sensitivity of retinal cells is not some fiction. Similarly tuning curves of neurons in v1, v2 etc. provide information -- need to be carefully interpreted of course.
Tuning curves have also given a lot of information about the hippocampus. Some of these expts are downright brilliant. Recently we were able to interpret these info into a unified view, but there is no way it could have happened without those experiments https://twitter.com/dileeplearning/status/1373191886907699203?s=20">https://twitter.com/dileeplea...
Again, tuning curves should be adequately contextualized and should not be taken literally as "therefore this neuron represents X"..but that is very different from thinking tuning curves do not provide any information. 15/23
So what would real neuroscientists have found about the microprocessor using their techniques? Some examples: Most voltages are 5 or 0, there is a clock, there is min pulse width, there is a bus, the bus clock is subsampled, some pins are interrupts...etc.
More importantly, I think neuroscientists would have identified the program as a manipulatable input (not a behavior as in the paper). They would then have written elementary programs that isolate specific functions. Load register A, Add B etc, gradually isolating the components.
Nobody is trying to reverse engineer ANNs -- we already know how to forward engineer them!
Visualizing neurons in ANNs is often for post-hoc story telling, not for hints on how to reverse-engineer them, very different from our situation with the brain. 18/23
Visualizing neurons in ANNs is often for post-hoc story telling, not for hints on how to reverse-engineer them, very different from our situation with the brain. 18/23
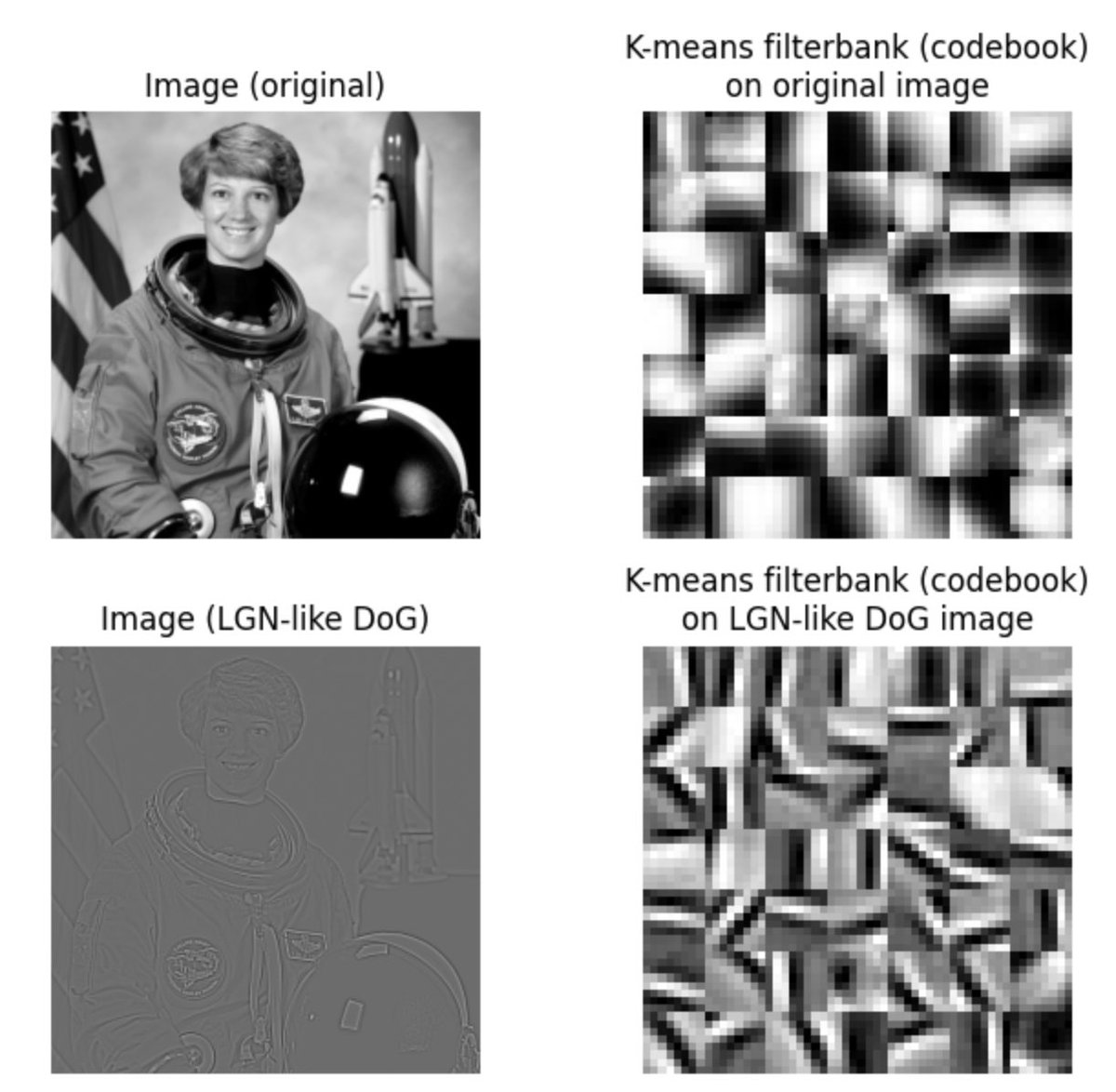
Here is the dictionary learned by k-means algorithm after running on an image. Do we "understand" how each of those filters came about? Does that mean we don& #39;t understand k-means? ...an example of the problem with the analogy above. 19/23
"There little reason to assume that any methods we used should be more meaningful on brains"...this is a strong claim. As I discussed above, there are many valid reasons for this being the case. 20/23
This doesn& #39;t mean there are no methodology problems in neuroscience. The paper was indeed successful in bringing attention to those. I think the microprocessor analogy is a useful tool provided it is used carefully, just like tools in neuroscience. 21/23
But contrary to the paper, I think a wealth of information already exists in neuroscience. These provide important, but partial, clues regarding the functioning of the system. Combined with graphical models, ML, and DL, these cues can be utilized to build theories and models. 22/
What the paper did not argue for, but I do:
1. More system-level, detailed, and buildable theories that pay attention to existing experimental data.
2. More hypothesis driven neuroscience experiments.
3. Tighter and faster iterations between 1&2.
/End.
1. More system-level, detailed, and buildable theories that pay attention to existing experimental data.
2. More hypothesis driven neuroscience experiments.
3. Tighter and faster iterations between 1&2.
/End.

 Read on Twitter
Read on Twitter