A new trans ethnic GWAS of type 2 diabetes has discovered an interesting locus (TGFB1; OR-1.27) with large effect size comparable to TCF7L2. The MAF of this variant is 0 in Europeans, but 6.8% in Africans. #appsec2">https://www.sciencedirect.com/science/article/pii/S2666247721000105?via%3Dihub #appsec2">https://www.sciencedirect.com/science/a...
It seems still there are many large effect size T2D loci waiting to be discovered, *but only through Non-Europeans*. Let me take this opportunity to revisit Vujovic et al, a GWAS of T2D in 1.4 million individuals, which came out last year.
https://www.nature.com/articles/s41588-020-0637-y.">https://www.nature.com/articles/...
https://www.nature.com/articles/s41588-020-0637-y.">https://www.nature.com/articles/...
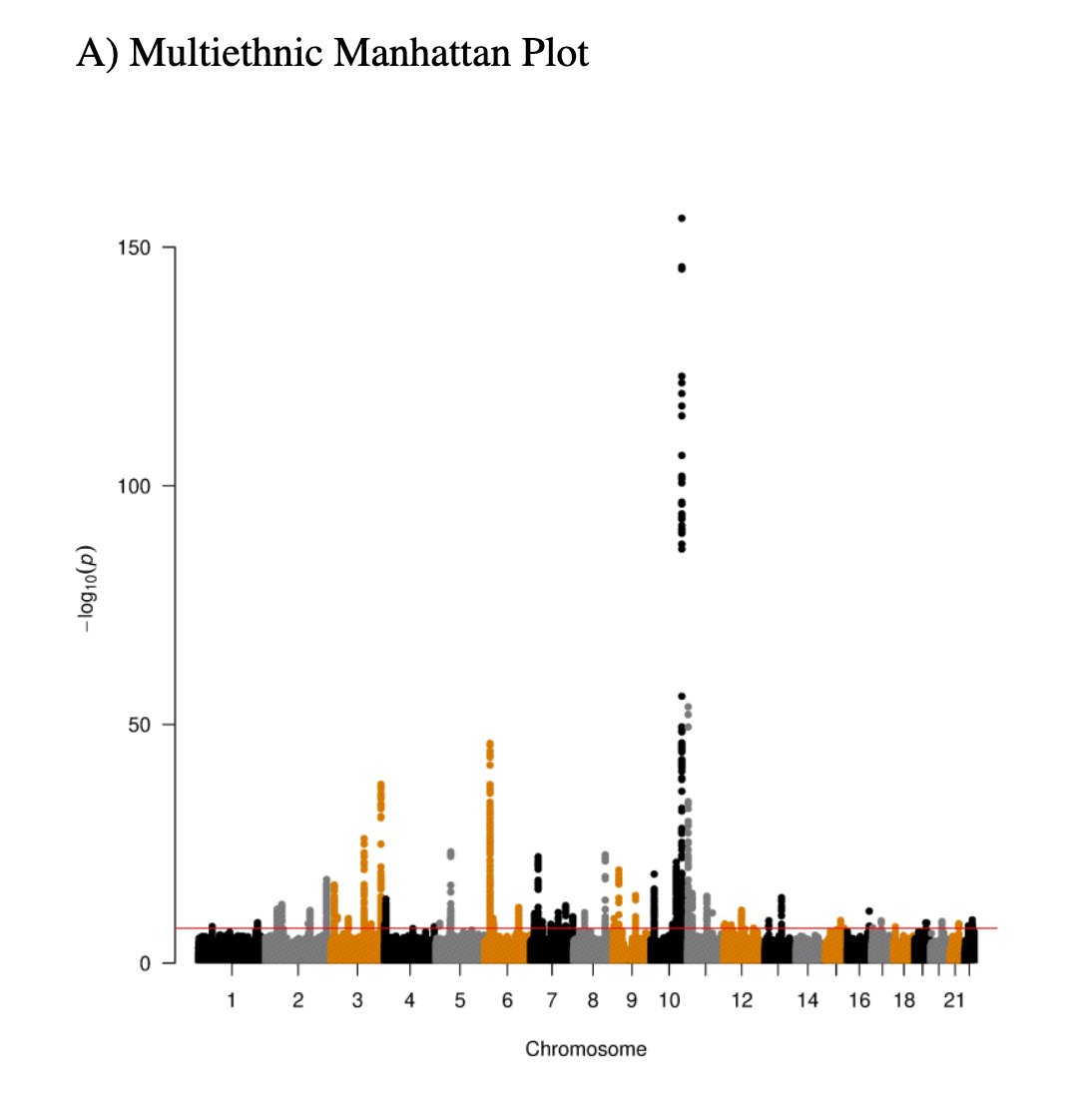
Let& #39;s see what kind of variants get newly discovered as the sample size grow bigger and bigger or when including individuals from non-European ancestries. I have illustrated this before for other traits, for e.g. telemore length. https://twitter.com/doctorveera/status/1375888339308544002?s=20">https://twitter.com/doctorvee...
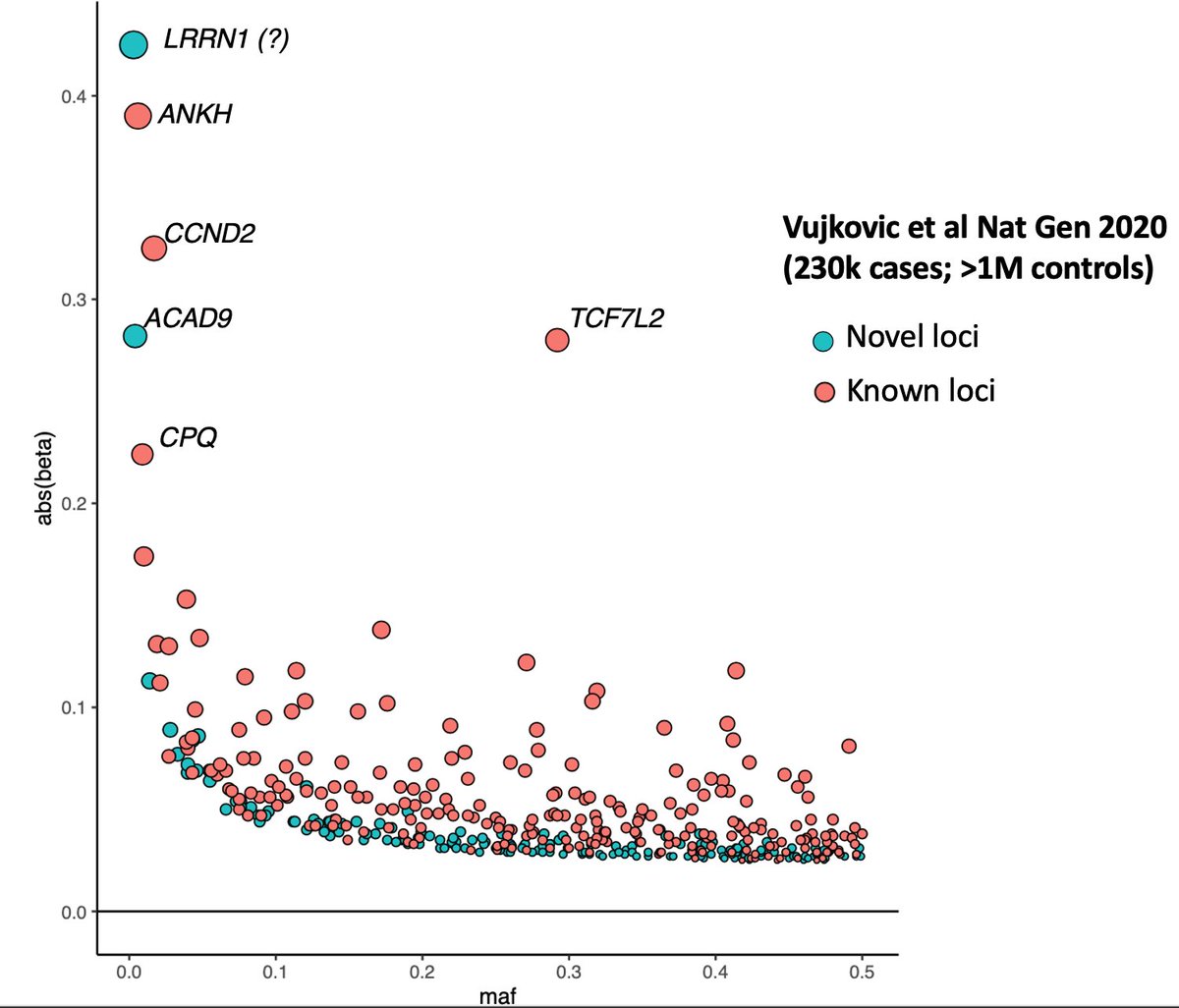
Let me illustrate using T2D as an example. Here in this plot I& #39;ve color coded the novel and known T2D loci (thanks to the authors who provided this as a neat supplementary table).
First thing to notice is that the most of the novel loci have lower effect sizes than known loci. This is rule 1: as N gets bigger and bigger, the effect sizes of the novel loci will be smaller and smaller. But this applies only to common variants.
So, if there are common variants in Europeans with large effect size we would have known by now. No matter how much we increase the N we will not find any more novel T2D common variants *in Europeans* with large effect size.
Also, let& #39;s take this opportunity to appreciate the TCF7L2 locus, which stands out of the crowd. One of the very first T2D loci discovered and one of the very few GWAS "candidate genes" that replicated again and again and again in almost every population. https://twitter.com/STRUANGRANT/status/1350572181344944129?s=20">https://twitter.com/STRUANGRA...
The second thing to notice in the plot is there are a couple of novel loci with large effect sizes, but they are rare in the Europeans, which means to discover them you need extremely large sample sizes.
This is rule 2: if the novel locus had a large effect size then it will be a rare variant. We have seen this now in two traits already T2D and telomere length.
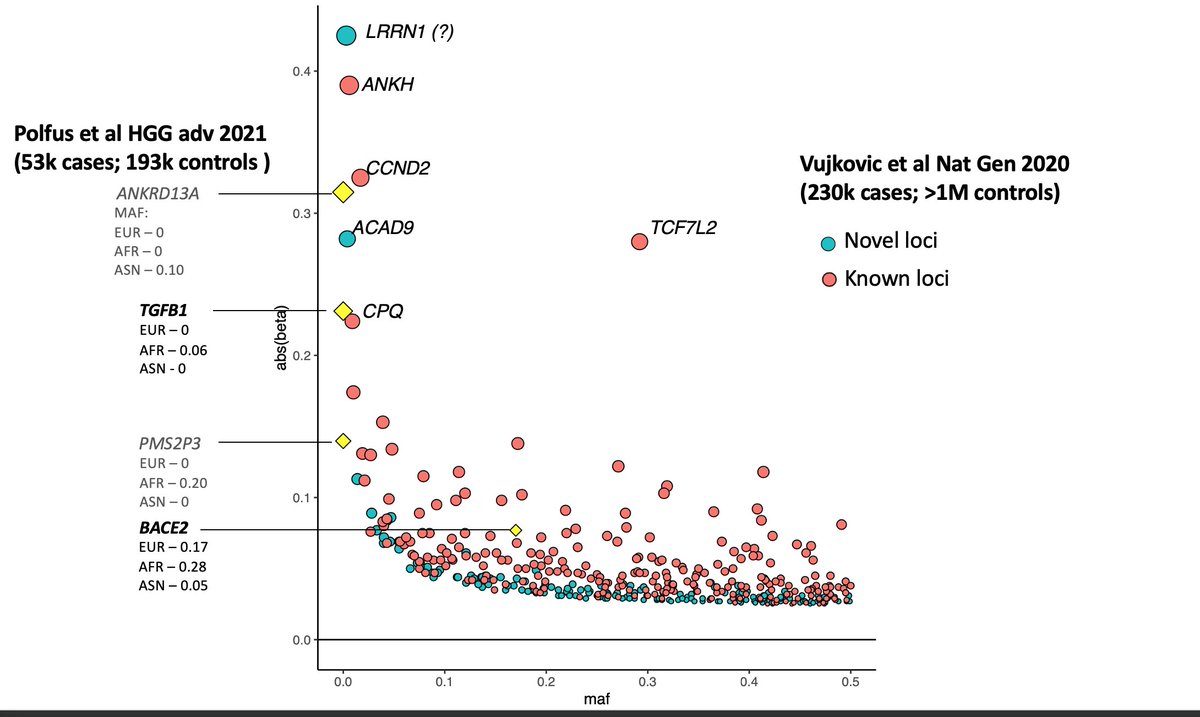
Now let& #39;s come to the current paper where the authors discovered four loci via trans ethnic GWAS, but were able to replicate only two (bold ones). I overlayed these four loci (yellow diamonds) on the previous plot.
Three of the four discovered loci have large effect sizes. Notice that all three are rare in Europeans. Should they have been common in Europeans, we would have discovered them by now.
This illustrates the advantage of studying non-Europeans. Consider TGFB1 locus, which was not discovered even after studying more than 1 million Europeans, but popped out in a GWAS of just a few thousand Africans (~8500 cases and ~16000 controls).
Large effect size loci will most often point us straight to the core pathways and possibly to drug targets. We had searched enough in the Europeans. To discover more T2D large effect locus, we should shift our focus towards the Non-Europeans. The current paper is a great example.
I should add that these interpretations apply only to variants detectable via arrays/imputation or exome sequencing. There is a special group of unexplored variants, large structural variants, which might turn out to have have huge effect size and common allele frequency.

 Read on Twitter
Read on Twitter