So this *finally* happened: https://twitter.com/TimWebster17/status/1331598518222524417?s=20">https://twitter.com/TimWebste...
And now, I present to you a thread about the first paper from my dissertation and (bonus milestone) my first ever preprint!
In this preprint, we describe new methods for quantifying hair fiber morphology.
"we" being @TimWebster17 @A_A_Zaidi , Nick Stephens, Kendall Routch and my advisors, Nina Jablonski & Mark Srhiver https://www.biorxiv.org/content/10.1101/2020.11.24.392191v1">https://www.biorxiv.org/content/1...
"we" being @TimWebster17 @A_A_Zaidi , Nick Stephens, Kendall Routch and my advisors, Nina Jablonski & Mark Srhiver https://www.biorxiv.org/content/10.1101/2020.11.24.392191v1">https://www.biorxiv.org/content/1...
Have you ever thought “oh, lemme just do this real quick”?
Well, that is how the story of this paper begins.
About 7 years ago, I dove down the rabbit hole that is measuring hair morphology.
I promise you, *nothing* about this was “real quick”
Well, that is how the story of this paper begins.
About 7 years ago, I dove down the rabbit hole that is measuring hair morphology.
I promise you, *nothing* about this was “real quick”
As tempting as it is to tell you the full story, I don’t want to write a 200-part Twitter thread.
So, suffice to say for background: measuring hair morphology is hard, people have sorta tried, but what was out there had issues so it didn’t really catch on
So, suffice to say for background: measuring hair morphology is hard, people have sorta tried, but what was out there had issues so it didn’t really catch on
What is hair morphology?
Usually people talk about “overall” hair morphology in terms of hair texture (e.g. straight, wavy, curly), but if you’re looking to measure things quantitatively at the level of the individual fiber, you can talk about 1) cross-section and 2) curvature
Usually people talk about “overall” hair morphology in terms of hair texture (e.g. straight, wavy, curly), but if you’re looking to measure things quantitatively at the level of the individual fiber, you can talk about 1) cross-section and 2) curvature
Cross-sectional morphology of the hair fiber is what you get when you slice a hair shaft in half (or more parts, if that’s your thing) and look at it under a microscope.
Generally, people talk about area and eccentricity.
Generally, people talk about area and eccentricity.
A note on eccentricity:
related terms include ellipticity, min/max diameter and I’ve even seen hair index
All these measures describe how close/far the cross-section is to a perfect circle. I like eccentricity because it goes from 0 to 1 (circular to elliptical)
related terms include ellipticity, min/max diameter and I’ve even seen hair index
All these measures describe how close/far the cross-section is to a perfect circle. I like eccentricity because it goes from 0 to 1 (circular to elliptical)
Curvature of a hair fiber refers to how tightly curled a hair fiber is.
If you try to fit a circle to a curve, you can take the inverse of its radius, so: the smaller or tighter the curl, the higher the curvature.
A straight hair has a radius of infinity and a curvature of 0
If you try to fit a circle to a curve, you can take the inverse of its radius, so: the smaller or tighter the curl, the higher the curvature.
A straight hair has a radius of infinity and a curvature of 0
So, if that’s it, what’s the big deal? Why aren’t there good methods to measure these aspects of hair?
Well, because hair is a horrible, finicky, demonic thing that will ruin your life. Both on your scalp and in the lab.
Jokes aside, sample prep is the main issue
Well, because hair is a horrible, finicky, demonic thing that will ruin your life. Both on your scalp and in the lab.
Jokes aside, sample prep is the main issue
To image the cross-section of a hair, you need to embed it and cut it.
Hair is disproportionately thin and long, so it’s nearly impossible to handle without encasing it in something so you can manipulate it more precisely (especially important if slicing because danger).
Hair is disproportionately thin and long, so it’s nearly impossible to handle without encasing it in something so you can manipulate it more precisely (especially important if slicing because danger).
But people have been embedding it in resin anyway. And lemme tell ya, not fun. That stuff takes like 24 hours to dry and you need hairs to lay parallel to each other to get a clean section.
“How do you keep a curly hair stretched straight for 24 hours?” I hear you ask…
“How do you keep a curly hair stretched straight for 24 hours?” I hear you ask…
Well, you can cut slits into a piece of cardboard or transparency and hook them into those.
But you will either end up with a massive block of resin that is a pain to slice…
But you will either end up with a massive block of resin that is a pain to slice…
...or, if you really hate yourself you can use the tiny molds the microscopy lab gives you and tweeze hairs into place just to find out they decided to start wilin out during the drying process :’)
Plus you need a microtome to get slices (expensive + hard to work with)
Plus you need a microtome to get slices (expensive + hard to work with)
After loads of detours, my advisor (Mark) and I just started playing around with random things in the lab (that’s science for ya) and he suggested we laminate the hairs
When I tell you I literally laughed out loud...
I was convinced there was no way it would work
Plot twist…
When I tell you I literally laughed out loud...
I was convinced there was no way it would work
Plot twist…
We tested it out and there was something about the plastic melting around the hairs that just really worked and it put us on the right track
Eventually, we settled on (polycaprolactone) plastic strips and here’s a video of what that process looks like: https://youtu.be/OSgiErHmiDE ">https://youtu.be/OSgiErHmi...
Eventually, we settled on (polycaprolactone) plastic strips and here’s a video of what that process looks like: https://youtu.be/OSgiErHmiDE ">https://youtu.be/OSgiErHmi...
Here is a link to the embedding and sectioning protocol we developed: https://dx.doi.org/10.17504/protocols.io.bbwcipaw">https://dx.doi.org/10.17504/...
The benefit of this over traditional resin + microtome method is that you can immediately embed hairs (vs. 24h drying) and you can cut them with a razor blade (much cheaper and easier than a microtome).
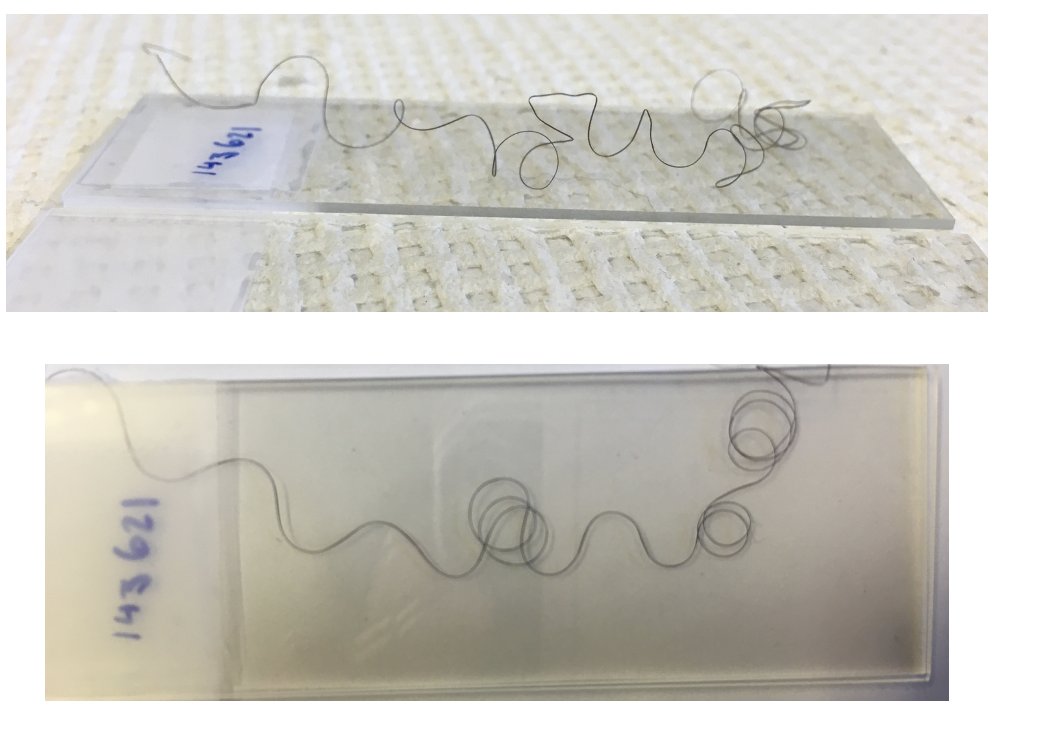
Then, all you have to do is mount vertically and image:
Then, all you have to do is mount vertically and image:
Once you have those images, you do a lil  https://abs.twimg.com/emoji/v2/... draggable="false" alt="✨" title="Funken" aria-label="Emoji: Funken">image analysis
https://abs.twimg.com/emoji/v2/... draggable="false" alt="✨" title="Funken" aria-label="Emoji: Funken">image analysis https://abs.twimg.com/emoji/v2/... draggable="false" alt="✨" title="Funken" aria-label="Emoji: Funken"> and bim bam boom, you got yourself some cross-sectional data.
https://abs.twimg.com/emoji/v2/... draggable="false" alt="✨" title="Funken" aria-label="Emoji: Funken"> and bim bam boom, you got yourself some cross-sectional data.
(Okay, there’s more to this, but here’s a flowchart of the program I wrote to do it)
(Okay, there’s more to this, but here’s a flowchart of the program I wrote to do it)
How do you prepare hair samples to study their curvature?
You don’t. You just turn around and find another subject of study so you don’t waste precious years of your life doing this random thing that no one asked you to do.
But, if you’re stubborn like me...
You don’t. You just turn around and find another subject of study so you don’t waste precious years of your life doing this random thing that no one asked you to do.
But, if you’re stubborn like me...
The main question is: how do you measure curvature in 2 dimensions when non-straight hair curls in 3 dimensions?
Previously, the solution was to flatten it between glass slides and use a transparency with arcs of known sizes to figure out the curl radii.
This is suboptimal.
Previously, the solution was to flatten it between glass slides and use a transparency with arcs of known sizes to figure out the curl radii.
This is suboptimal.
Not only does it take forever to do, flattening the hair also distorts the hair’s shape in a way that is unpredictable.
Also, what do you do about hairs of different lengths? Is it really comparable? And how do you wash the hairs?
So, we came up with a solution
Also, what do you do about hairs of different lengths? Is it really comparable? And how do you wash the hairs?
So, we came up with a solution
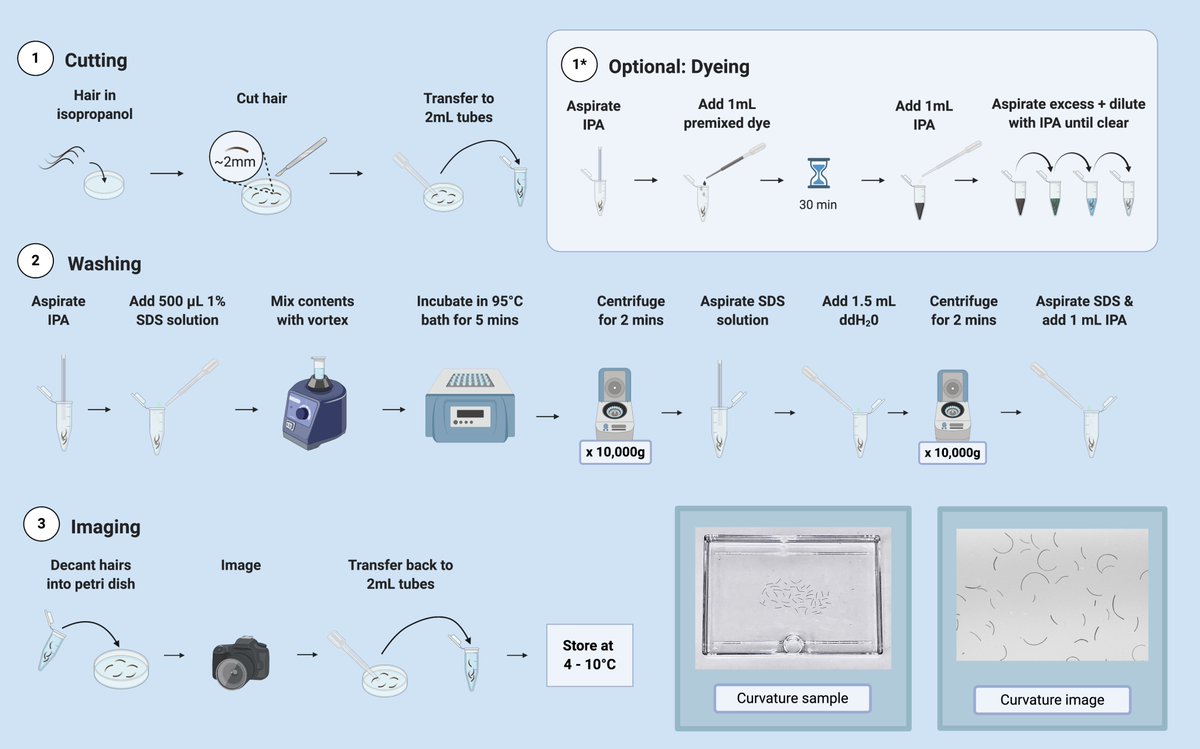
First, it was important to find a way to wash the hairs that wasn’t tweezing each hair fiber in and out of boiling water (I did it - would not recommend)
We ended up developing a protocol that cut the hairs to uniform length (~3mm) and washed it in a way that was scalable
We ended up developing a protocol that cut the hairs to uniform length (~3mm) and washed it in a way that was scalable
All the diagrams were made with @BioRender btw
Also there’s this video of our methods: https://youtu.be/qR-Jj4K0Bts ">https://youtu.be/qR-Jj4K0B...
Also there’s this video of our methods: https://youtu.be/qR-Jj4K0Bts ">https://youtu.be/qR-Jj4K0B...
And the full protocol is here on @protocolsIO https://dx.doi.org/10.17504/protocols.io.bbweipbe">https://dx.doi.org/10.17504/...
Developing methods for this was a whole adventure. Especially when sifting through some of the more ancient pieces of equipment in the lab. However, I can confirm, a Vortex mixer from the 80’s will likely outlive you: https://twitter.com/Prokaryota/status/1331553218573156358">https://twitter.com/Prokaryot...
The last part of our methods is a Python package that I developed with the help of @TimWebster17 and Nick Stephens (a postdoc w/ @TimRyanAnth at Penn State).
You can find it on PyPi: https://pypi.org/project/fibermorph/">https://pypi.org/project/f...
and GitHub: https://github.com/tinalasisi/fibermorph">https://github.com/tinalasis...
You can find it on PyPi: https://pypi.org/project/fibermorph/">https://pypi.org/project/f...
and GitHub: https://github.com/tinalasisi/fibermorph">https://github.com/tinalasis...
I spent a ridiculous amount of time writing this program because I have never taken a single programming course in my life and had no idea what I was doing when I started out.
No joke, I was a noob: a year ago, I was calling the terminal The Black Screen™
No joke, I was a noob: a year ago, I was calling the terminal The Black Screen™
It was important to me that what I made was actually *usable* to people who weren’t programmers.
So, our program runs on the command line and requires no coding skills, just a folder where your images live, basically
So, our program runs on the command line and requires no coding skills, just a folder where your images live, basically
Now, the program is by no means perfect, but it works and it& #39;s in a state where others can actually take on certain parts of it and improve them
(Someone please go through my code and make unit tests for all my functions, thank you)
(Someone please go through my code and make unit tests for all my functions, thank you)
The two hardest lessons to learn with the code were:
1) If it works, it works. At some point, you gotta let it out into the world
2) Just cause someone says they’re great at coding, doesn’t mean they are https://twitter.com/dangerouswco/status/1326526035186343936?s=20">https://twitter.com/dangerous...
1) If it works, it works. At some point, you gotta let it out into the world
2) Just cause someone says they’re great at coding, doesn’t mean they are https://twitter.com/dangerouswco/status/1326526035186343936?s=20">https://twitter.com/dangerous...
Last coding related tangent:
I’m infinitely grateful to @TimWebster17 and Nick for guiding me through the process. It’s opened a whole new world for me.
I never thought I’d be able to code because I wasn’t “good at math”
Turns out, I *can* learn and I actually love it!
I’m infinitely grateful to @TimWebster17 and Nick for guiding me through the process. It’s opened a whole new world for me.
I never thought I’d be able to code because I wasn’t “good at math”
Turns out, I *can* learn and I actually love it!
Now, onto our results.
The main question is: is measuring hair fiber morphology quantitatively worth it?
… Yes
(bet you didn’t see that coming, huh?)
The main question is: is measuring hair fiber morphology quantitatively worth it?
… Yes
(bet you didn’t see that coming, huh?)
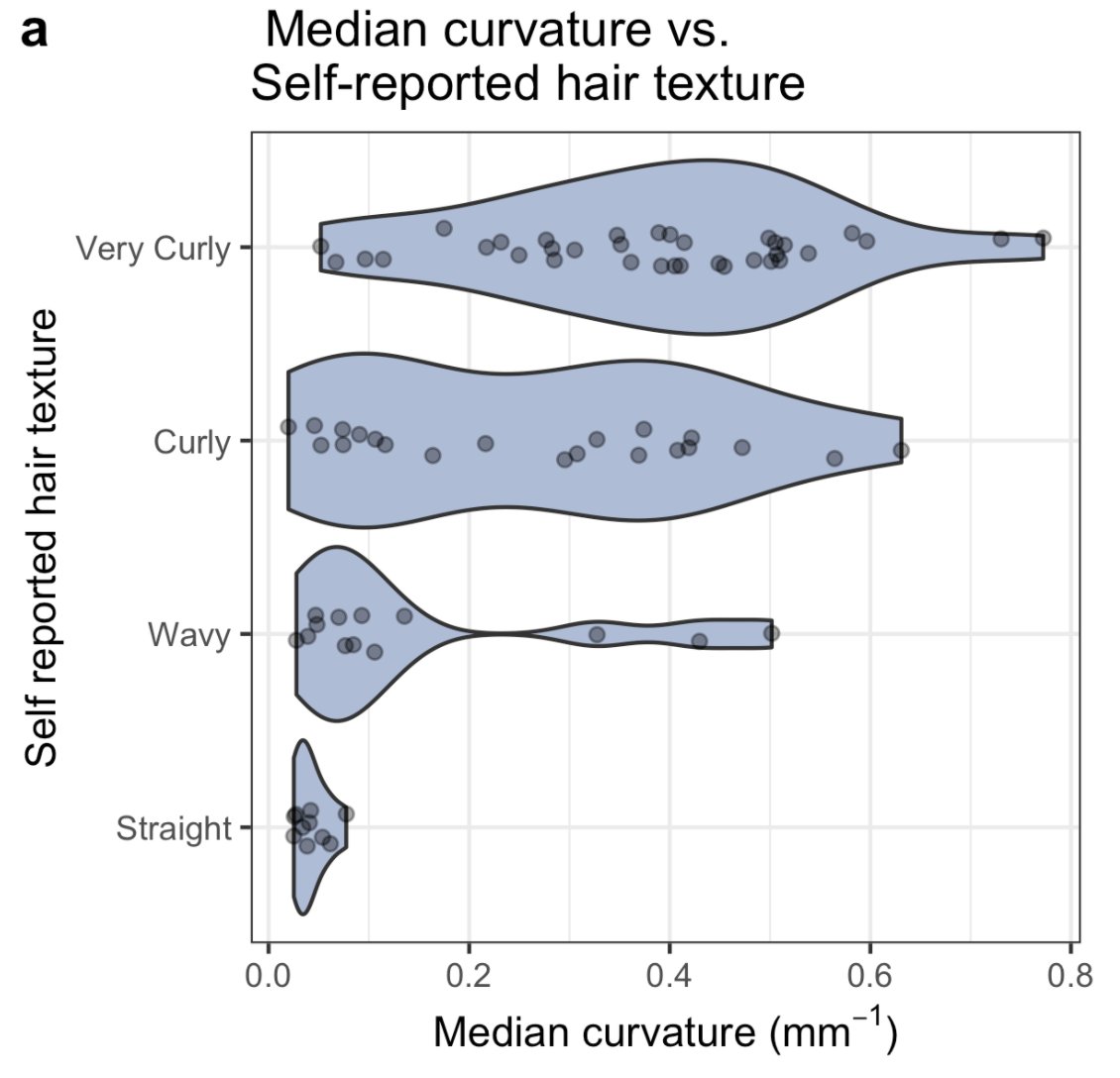
If you compare self-reported hair texture to quantitative curvature, you see that there seems to be a bit of a distribution in terms of what people think the different categories mean:
There’s some literature out there that suggests you can classify hairs into categories based on a curvature threshold (among other variables), but what we see there is that the cut offs they suggest are heavily biased towards distinguishing low-curvature hair morphology
My biggest frustration about all these commonly used classification schemes is that they are ridiculously Eurocentric.
The number of times I’ve seen people distinguish between “wavy” and “curly” hair but lump together all “frizzy” hair is too many.
But moving on...
The number of times I’ve seen people distinguish between “wavy” and “curly” hair but lump together all “frizzy” hair is too many.
But moving on...
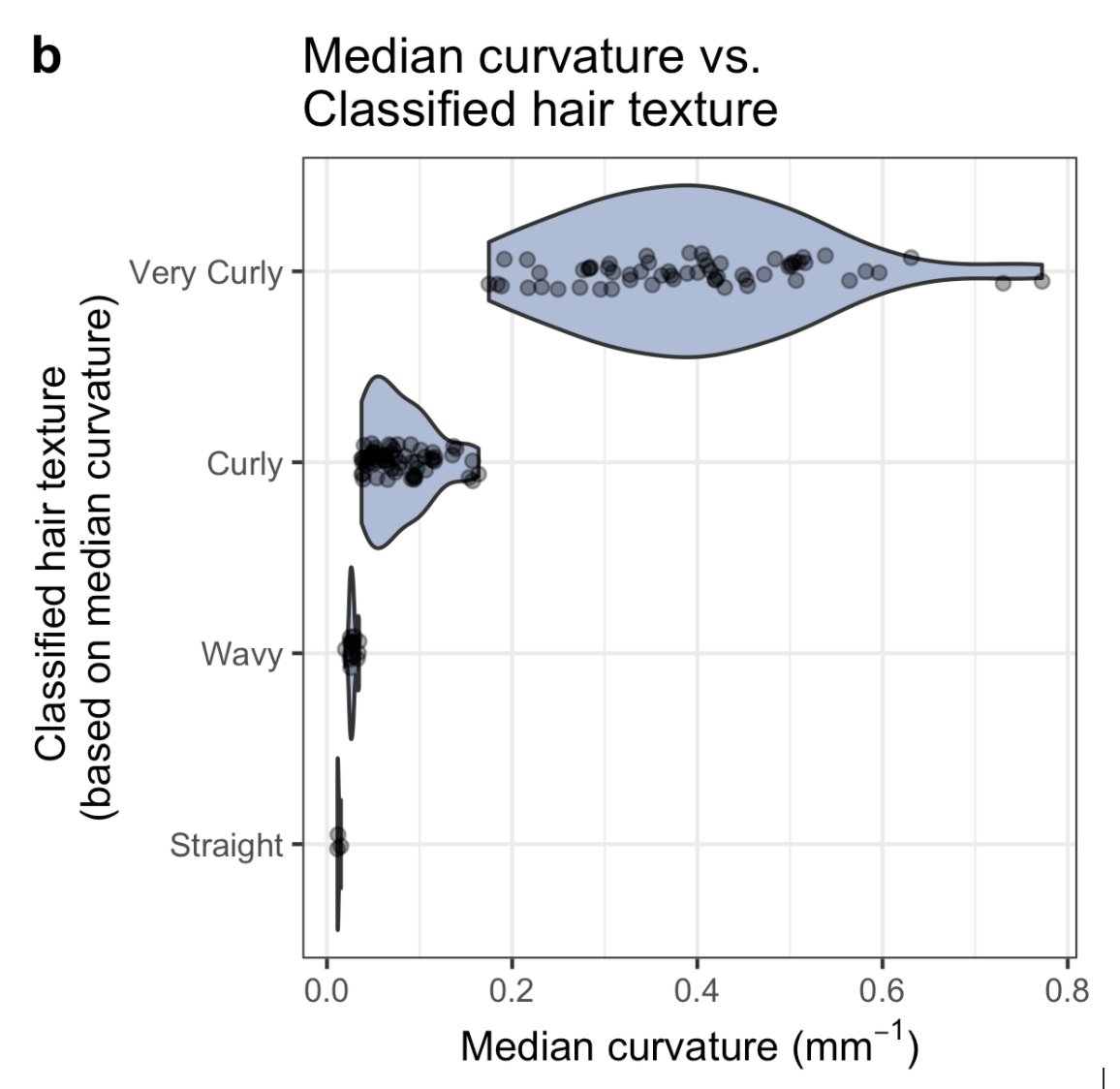
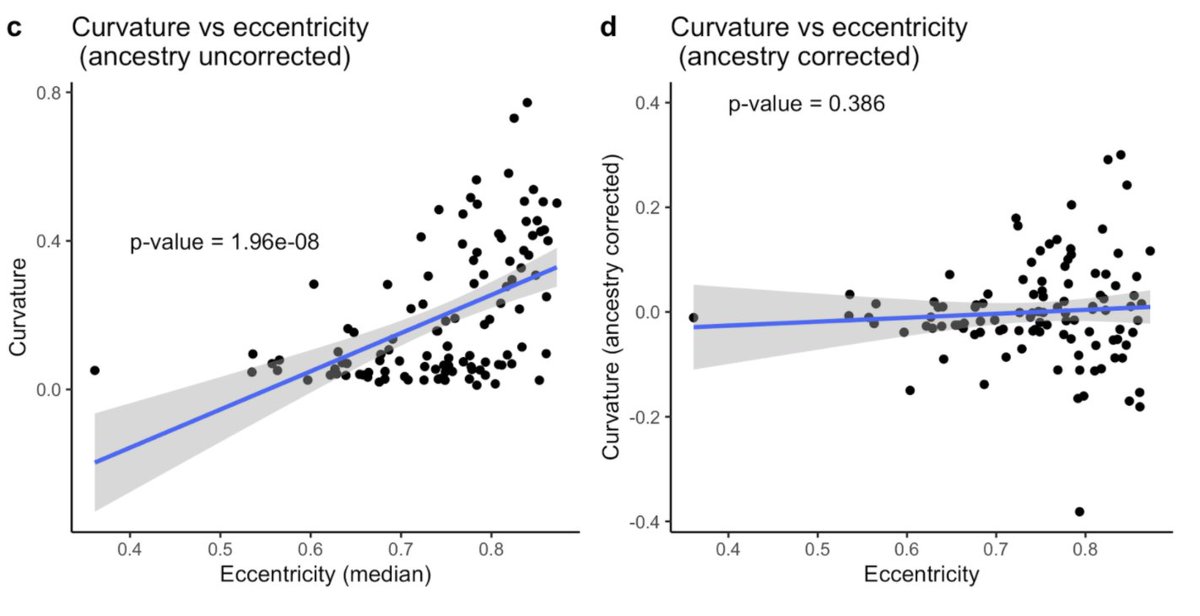
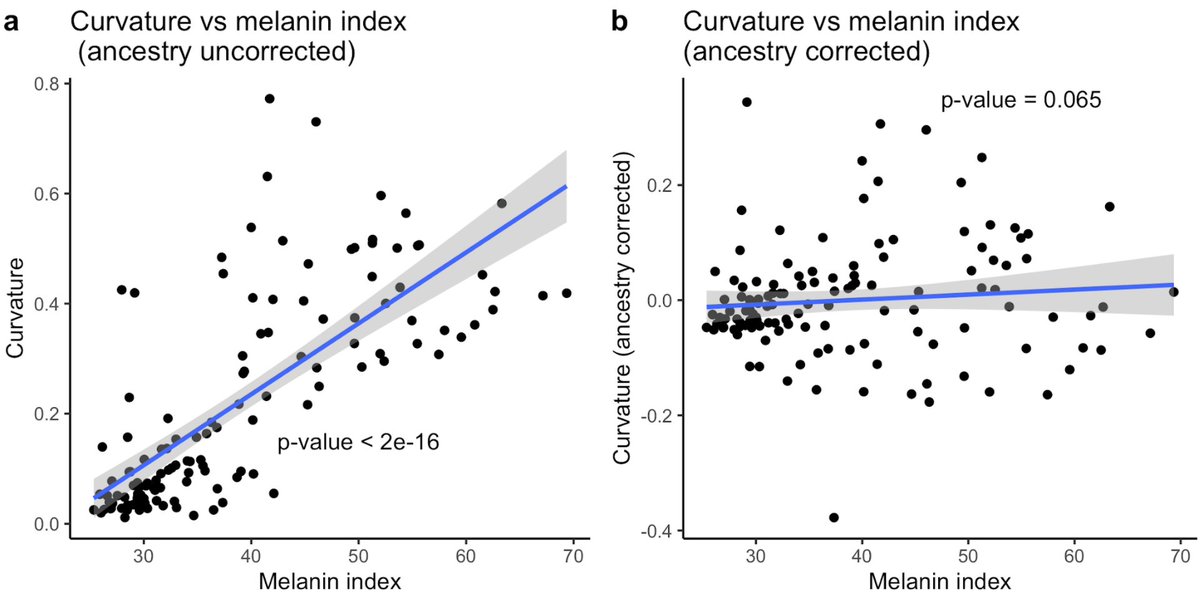
Okay, quantitative hair morphology shows more than qualitative, but is that variation meaningful?
Admixed populations can be a useful tool for studying traits that vary between populations.
They can be used to disentangle the effects of genetic and environmental variation
Admixed populations can be a useful tool for studying traits that vary between populations.
They can be used to disentangle the effects of genetic and environmental variation
A correlation with genetic ancestry can give you an idea of how heritable the difference between populations is.
We show that the correlation between curvature and ancestry in a sample of 140 people of mixed African & European ancestry is 85%
We show that the correlation between curvature and ancestry in a sample of 140 people of mixed African & European ancestry is 85%
For comparison, we looked at skin pigmentation (a trait that’s better understood):
In our sample, there was an 83% correlation between ancestry & skin pigmentation.
This pattern is *really* encouraging for future GWAS deciding to quantify this trait
In our sample, there was an 83% correlation between ancestry & skin pigmentation.
This pattern is *really* encouraging for future GWAS deciding to quantify this trait
One myth we were able to bust with our study is the idea that cross-sectional shape dictates hair curvature.
When you take into account ancestry, there is no correlation. So there probably isn’t a physical link between how “flat” a hair is and how curved it is.
When you take into account ancestry, there is no correlation. So there probably isn’t a physical link between how “flat” a hair is and how curved it is.
The correlation is a product of population structure, similar to what you see comparing hair curvature and skin pigmentation.
There’s no reason to believe these traits are biologically linked and once you take into account ancestry, that’s clear
There’s no reason to believe these traits are biologically linked and once you take into account ancestry, that’s clear
If you want to check out our stuff, here are the relevant links:
PyPi page for the fibermorph package: https://pypi.org/project/fibermorph/">https://pypi.org/project/f...
PyPi page for the fibermorph package: https://pypi.org/project/fibermorph/">https://pypi.org/project/f...
There’s a project website I made with workflowr: https://tinalasisi.github.io/2020_HairPheno_manuscript/index.html
(Thank">https://tinalasisi.github.io/2020_Hair... you @jdblischak !)
(Thank">https://tinalasisi.github.io/2020_Hair... you @jdblischak !)
And the raw images we used are on Zenodo: https://doi.org/10.5281/zenodo.4289252">https://doi.org/10.5281/z...
So, that’s it for our new methods. I hope they will be useful to folks across disciplines who study hair morphology.
Everything is open source and open access, so I hope folks can take on whatever part of the project strikes their fancy (lab or computational) and improve on that
Everything is open source and open access, so I hope folks can take on whatever part of the project strikes their fancy (lab or computational) and improve on that
Developing these methods was a gargantuan task. Not exaggerating even a little. At age 28, I shouldn’t be able to say I did anything for 7 years (¼ of my life, 100% of my adult life)
This also wouldn’t have been possible without the dozens of people who helped me along the way
This also wouldn’t have been possible without the dozens of people who helped me along the way
First a huge shout out to all the undergrads who helped me test out various methods and a special thank you to Duneshka Cruz and Sam Muller who filmed and edited the videos for the protocol
Also check out Sam’s art: https://www.instagram.com/paintedanatomy/ ">https://www.instagram.com/paintedan...
Also check out Sam’s art: https://www.instagram.com/paintedanatomy/ ">https://www.instagram.com/paintedan...
Thank you to Kendall Routch (one of the co-authors) who was an undergraduate research assistant in our lab.
She took charge of testing out all the variations of our embedding protocol and then took charge of embedding and imaging our samples.
She took charge of testing out all the variations of our embedding protocol and then took charge of embedding and imaging our samples.
And a huge thank you to @A_A_Zaidi who has been my friend for years but also became my full-fledged mentor during this project. Not only did he make analyses fun (no cap, we actually have fun Zoom work sessions), but he’s also made me feel genuinely supported.
Finally, @TimWebster17 : I genuinely don’t remember how I ended up roping you into this, but thank you for helping me write a legit Python package(!!!) and serving as a life coach when I thought this project would literally never end
And that’s all for now, I have another deadline to deal with  https://abs.twimg.com/emoji/v2/... draggable="false" alt="😬" title="Grimasse schneidendes Gesicht" aria-label="Emoji: Grimasse schneidendes Gesicht">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="😬" title="Grimasse schneidendes Gesicht" aria-label="Emoji: Grimasse schneidendes Gesicht">
Lol, remember when I said I didn& #39;t want to make this thread too long?  https://abs.twimg.com/emoji/v2/... draggable="false" alt="🤡" title="Clownsgesicht" aria-label="Emoji: Clownsgesicht"> https://twitter.com/TinaLasisi/status/1331681854966075396?s=20">https://twitter.com/TinaLasis...
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🤡" title="Clownsgesicht" aria-label="Emoji: Clownsgesicht"> https://twitter.com/TinaLasisi/status/1331681854966075396?s=20">https://twitter.com/TinaLasis...

 Read on Twitter
Read on Twitter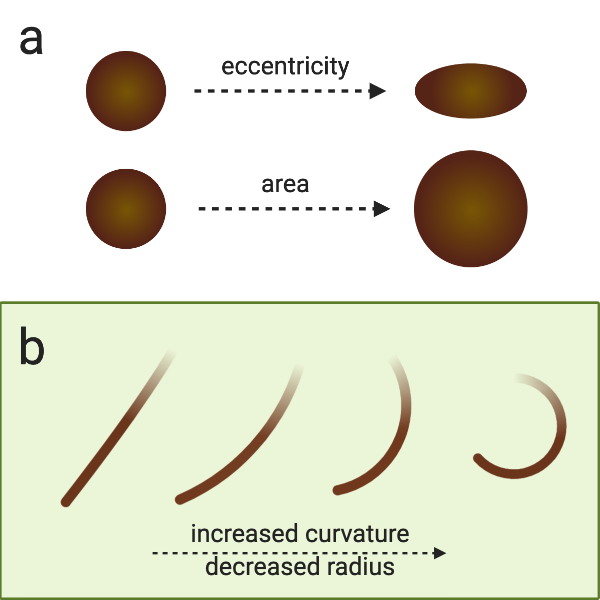
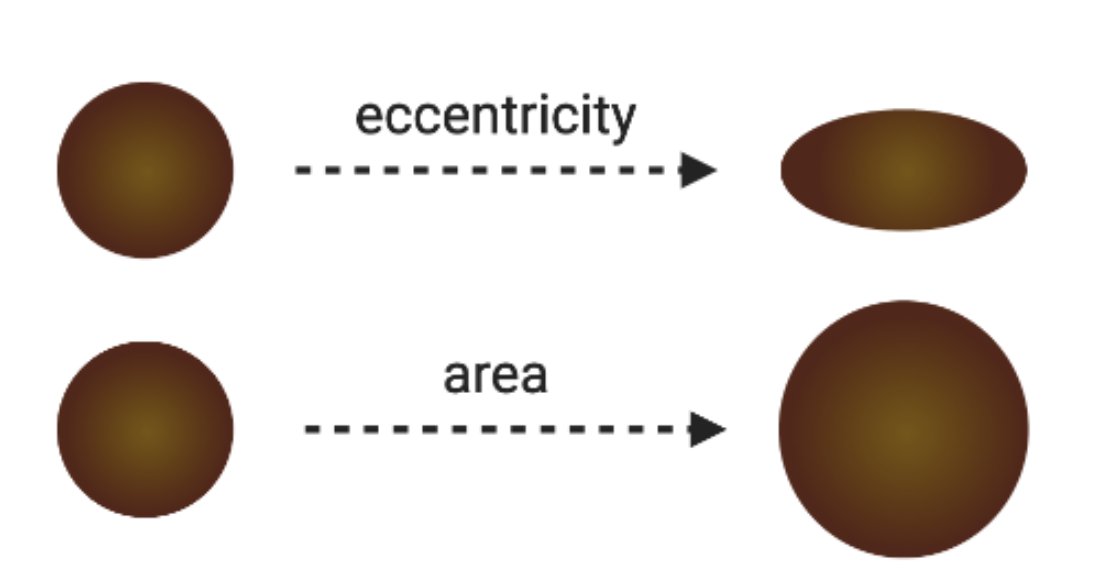
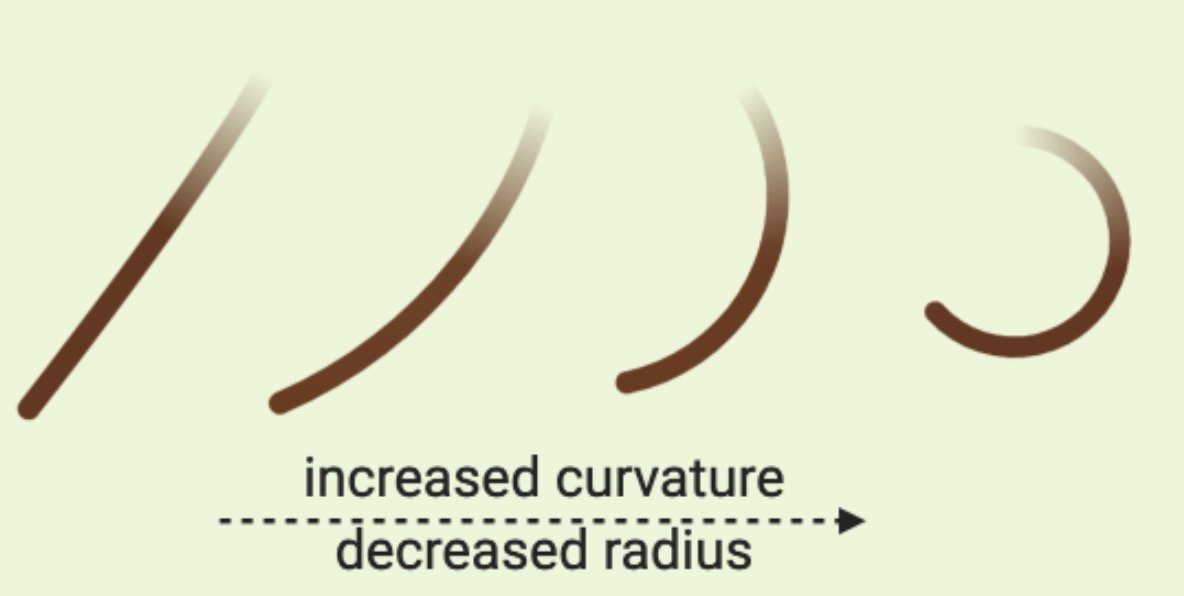



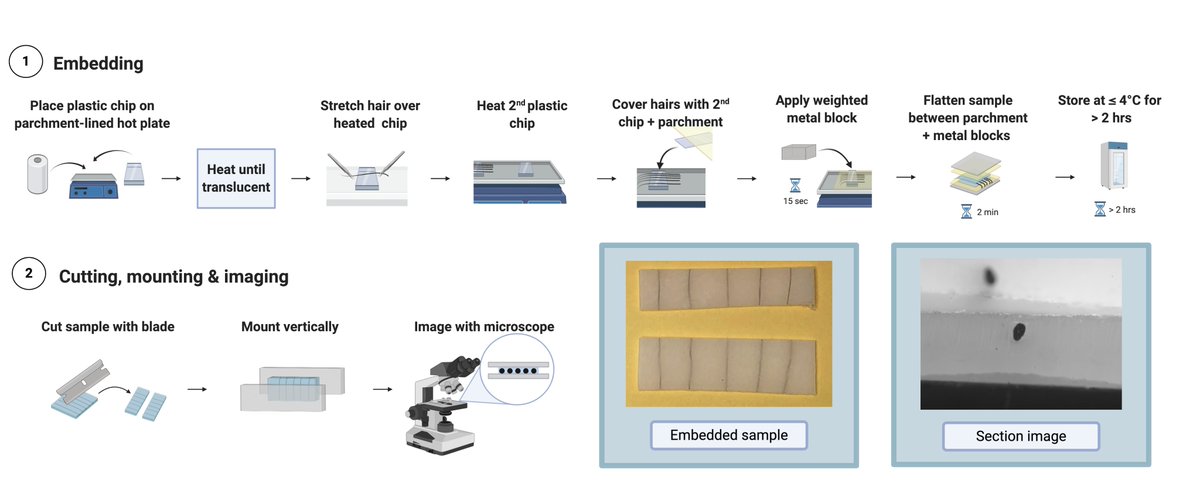
 image analysishttps://abs.twimg.com/emoji/v2/... draggable="false" alt="✨" title="Funken" aria-label="Emoji: Funken"> and bim bam boom, you got yourself some cross-sectional data.(Okay, there’s more to this, but here’s a flowchart of the program I wrote to do it)" title="Once you have those images, you do a lil https://abs.twimg.com/emoji/v2/... draggable="false" alt="✨" title="Funken" aria-label="Emoji: Funken">image analysishttps://abs.twimg.com/emoji/v2/... draggable="false" alt="✨" title="Funken" aria-label="Emoji: Funken"> and bim bam boom, you got yourself some cross-sectional data.(Okay, there’s more to this, but here’s a flowchart of the program I wrote to do it)" class="img-responsive" style="max-width:100%;"/>
image analysishttps://abs.twimg.com/emoji/v2/... draggable="false" alt="✨" title="Funken" aria-label="Emoji: Funken"> and bim bam boom, you got yourself some cross-sectional data.(Okay, there’s more to this, but here’s a flowchart of the program I wrote to do it)" title="Once you have those images, you do a lil https://abs.twimg.com/emoji/v2/... draggable="false" alt="✨" title="Funken" aria-label="Emoji: Funken">image analysishttps://abs.twimg.com/emoji/v2/... draggable="false" alt="✨" title="Funken" aria-label="Emoji: Funken"> and bim bam boom, you got yourself some cross-sectional data.(Okay, there’s more to this, but here’s a flowchart of the program I wrote to do it)" class="img-responsive" style="max-width:100%;"/>