Paper out in Nature Comms. We developed a framework to analyse the contribution of #SARSCoV2 mutations to the virus& #39; transmissibility. We applied it to nearly 50k genomes and we found none (zero, zilch, nada) that increases transmission!
1/
https://www.nature.com/articles/s41467-020-19818-2">https://www.nature.com/articles/...
1/
https://www.nature.com/articles/s41467-020-19818-2">https://www.nature.com/articles/...
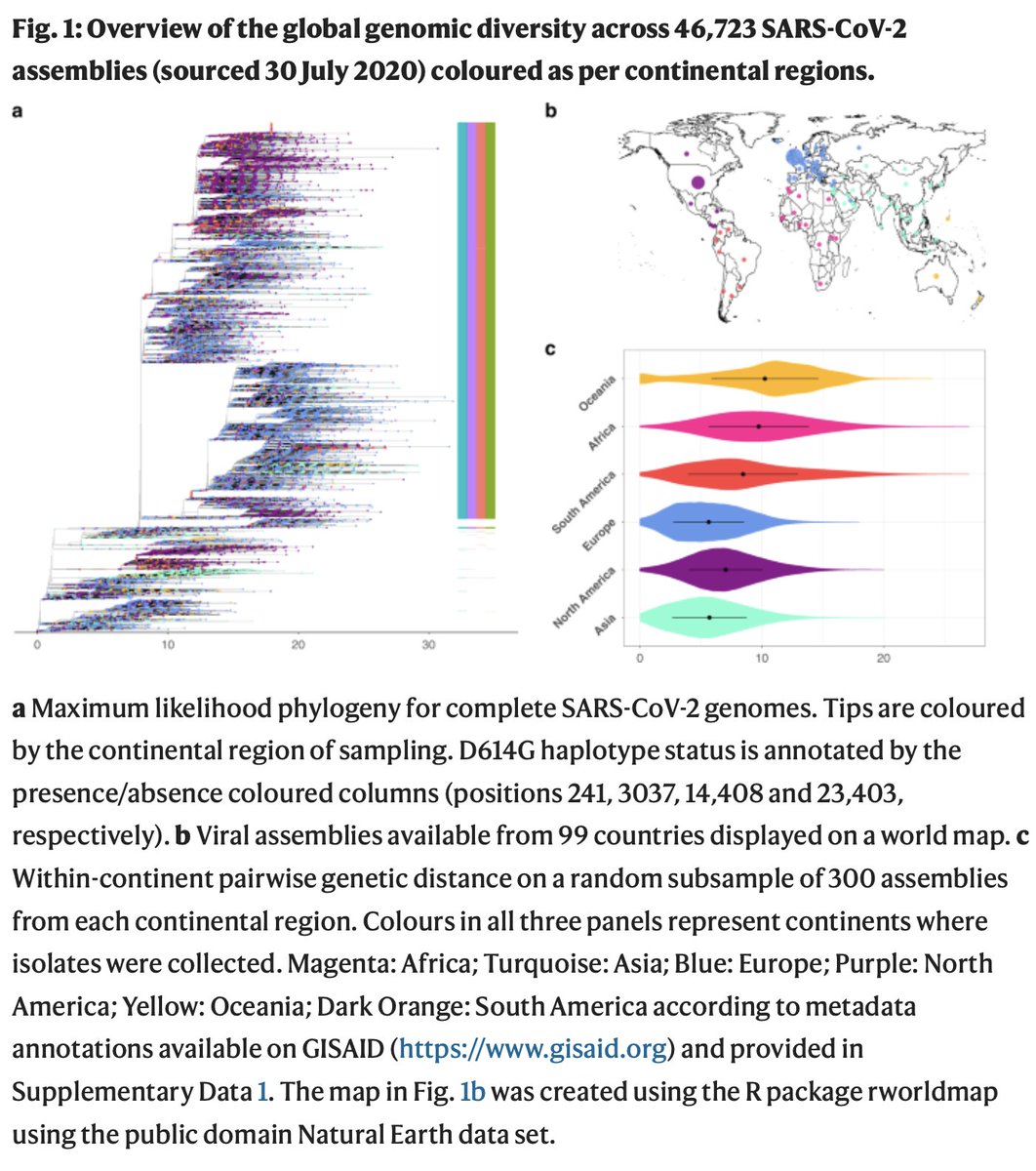
We identified 12,706 mutations in #SARSCoV2. 398 of those occurred repeatedly and independently. We honed on the 185 mutations which have occurred at least three times independently during the course of the pandemic.
2/ https://www.nature.com/articles/s41467-020-19818-2">https://www.nature.com/articles/...
2/ https://www.nature.com/articles/s41467-020-19818-2">https://www.nature.com/articles/...
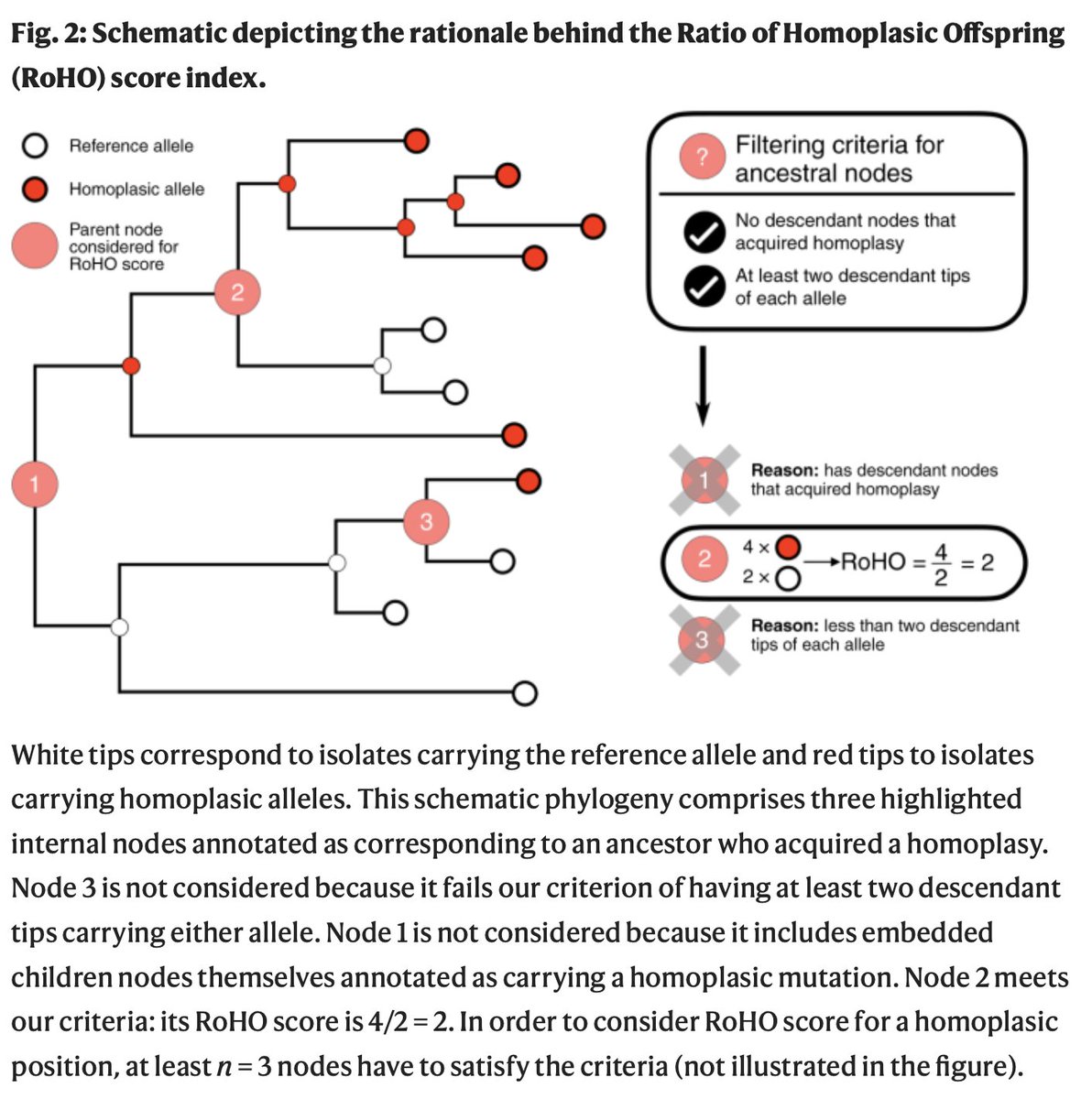
To test if mutations increase transmission of the virus carrying them, we modelled whether, after a mutation emerges, descendants of that virus outperform sister lineages without that particular mutation.
3/
https://www.nature.com/articles/s41467-020-19818-2">https://www.nature.com/articles/...
3/
https://www.nature.com/articles/s41467-020-19818-2">https://www.nature.com/articles/...
Mutations that are fairly common all seem neutral for the virus carrying them. This includes D614G, which according to our analysis is more of a stowaway that got a lucky ride on a successful lineage, rather than a driver of transmission.
4/
https://www.nature.com/articles/s41467-020-19818-2">https://www.nature.com/articles/...
4/
https://www.nature.com/articles/s41467-020-19818-2">https://www.nature.com/articles/...
This raises the question why #SARSCoV2 is so well adapted for transmission in humans. A plausible answer is that we missed the early window when it adapted to humans. For more, and about what happened when it later jumped into minks, see:
5/ https://twitter.com/BallouxFrancois/status/1328619165410799619">https://twitter.com/BallouxFr...
5/ https://twitter.com/BallouxFrancois/status/1328619165410799619">https://twitter.com/BallouxFr...
So, did we waste our time developing and optimising a framework to identify mutations that increase viral transmission, and that detected none? Not necessarily, the imminent arrival of vaccines will exert new selective pressures on #SARSCoV2.
6/ https://twitter.com/BallouxFrancois/status/1331042659230773248">https://twitter.com/BallouxFr...
6/ https://twitter.com/BallouxFrancois/status/1331042659230773248">https://twitter.com/BallouxFr...
mutations helping #SARSCoV2 to escape & #39;vaccine-induced immunity& #39; will allow viral lineages carrying them to transmit better. Our framework is well suited to flag such & #39;vaccine-escape& #39; mutations early, which should be helpful to inform vaccine updates.
7/ https://www.nature.com/articles/s41467-020-19818-2">https://www.nature.com/articles/...
7/ https://www.nature.com/articles/s41467-020-19818-2">https://www.nature.com/articles/...
The paper includes other results including on the critical role of the human immune system in inducing specific mutations in the virus, as well as an analysis of the amount of genetic recombination of the #SARSCoV2 population.
8/ https://www.nature.com/articles/s41467-020-19818-2">https://www.nature.com/articles/...
8/ https://www.nature.com/articles/s41467-020-19818-2">https://www.nature.com/articles/...
This paper represented hundreds of hours of work by a wonderful team including postdocs, a PhD student, and an undergraduate student ... and it cost essentially nothing to the taxpayer as we had no funding for it.
9/ https://www.nature.com/articles/s41467-020-19818-2">https://www.nature.com/articles/...
9/ https://www.nature.com/articles/s41467-020-19818-2">https://www.nature.com/articles/...
Thank you @LucyvanDorp for being the most wonderful colleague, and the rest of the team, Damien Richard, @CedricCSTan1, Liam Shaw and @misac42 for your dedication, resourcefulness, extraordinary hard work and for being such amazing collaborators.
10/ https://www.nature.com/articles/s41467-020-19818-2">https://www.nature.com/articles/...
10/ https://www.nature.com/articles/s41467-020-19818-2">https://www.nature.com/articles/...

 Read on Twitter
Read on Twitter