Hi Science Twitter!
A big shout out to my two fantastic colleagues Young Mi and Kevin, who have just released what is likely the largest tumour phylogeny built to date: the tree of a SINGLE cancer, recreated from >600 biopsies! A thread (1/10)
 https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Rückhand Zeigefinger nach unten" aria-label="Emoji: Rückhand Zeigefinger nach unten">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Rückhand Zeigefinger nach unten" aria-label="Emoji: Rückhand Zeigefinger nach unten">
https://doi.org/10.1371/journal.pbio.3000926">https://doi.org/10.1371/j...
A big shout out to my two fantastic colleagues Young Mi and Kevin, who have just released what is likely the largest tumour phylogeny built to date: the tree of a SINGLE cancer, recreated from >600 biopsies! A thread (1/10)
https://doi.org/10.1371/journal.pbio.3000926">https://doi.org/10.1371/j...
This transmissible cancer, devil facial tumour disease 1 (DFT1), has decimated Tasmanian devil populations for nearly ~25 years. Uncounted hours of fieldwork by many amazing collaborators in Tasmania led to this huge sample collection over the course of ~15 years. (2/10)
By sequencing whole DFT1 genomes at low coverage (~1X), this copy-number based tree provides fascinating insights into the disease origin and transmission across the island. For example, an early split between four major clades is followed by their geograpical separation. (3/10)
Only a single biopsy – likely corresponding to an extinct, ancestral clade – lacks a copy number loss that affects the beta-2 microglobulin (B2M) gene. This indicates an early adaptation in the tumours& #39; immune evasion strategy upon transmission events between devils. (4/10)
Already in 2013, @SiddleHannah, @KathyBelov, @Yuanyuan929 et al. found that B2M and MHC I are downregulated in DFT1 tumours ( https://www.pnas.org/content/110/13/5103).">https://www.pnas.org/content/1... In 2018, we noticed the B2M copy loss in DFT1 cell lines ( https://doi.org/10.1016/j.ccell.2018.03.013)">https://doi.org/10.1016/j... – now we can "date" this key mutation. (5/10)
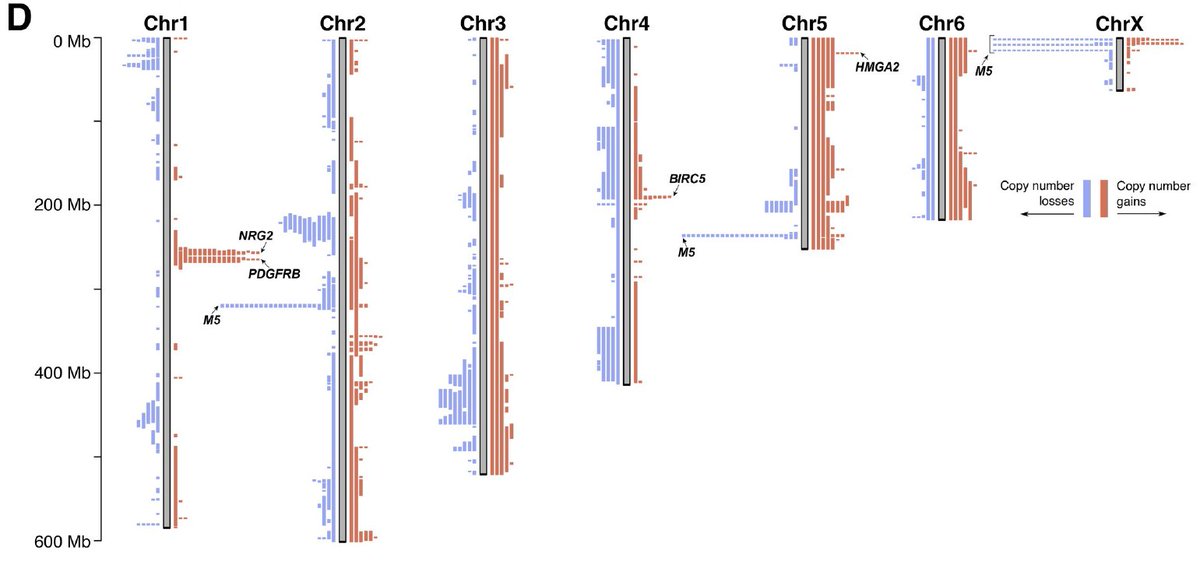
Recurrent copy-number changes also indicate other likely functional events: for example, the PDGFRB, BIRC5 and HMGA2 genes undergo frequent amplification in different branches of the tree. From studies in humans, we know that all three can be involved in oncogenesis. (6/10)
Beware – we also see that cell line cultivations do lead to genomic alterations in DFT1. Significantly increased copy-number aberrations hint at adaptations to cell culture conditions, irrespective of the source biopsy& #39;s phylogenetic positioning in the tree! (7/10)
What& #39;s more is that these cell culture-associated CNVs tend to be highly recurrent: nearly all the 24 lines sequenced feature a loss of one arm of Chr4 (incl. ERBB2) and a concomitant gain of one arm of Chr5. Wild stuff. (8/10)
Read the full story in @PLOSBiology for further insights, such as on regional lineage replacements, co-infections, marker chromosome formation and losses, fragile genome sites, telomeres and many more ... (9/10)
Super happy to be part of this study over the last ~5 years! Thanks to our mentor Liz Murchison, wonderful colleagues and friends at UTAS/STDP/Menzies, Kevin, Young Mi and I could even join for a number of devil monitoring adventures in Tasmania ourselves https://abs.twimg.com/emoji/v2/... draggable="false" alt="🐾" title="Pfotenabdrücke" aria-label="Emoji: Pfotenabdrücke">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🐾" title="Pfotenabdrücke" aria-label="Emoji: Pfotenabdrücke"> https://abs.twimg.com/emoji/v2/... draggable="false" alt="🥾" title="Hiking boot" aria-label="Emoji: Hiking boot">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🥾" title="Hiking boot" aria-label="Emoji: Hiking boot"> https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌏" title="Asien-Australien auf dem Globus" aria-label="Emoji: Asien-Australien auf dem Globus">(10/10)
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌏" title="Asien-Australien auf dem Globus" aria-label="Emoji: Asien-Australien auf dem Globus">(10/10)

 Read on Twitter
Read on Twitter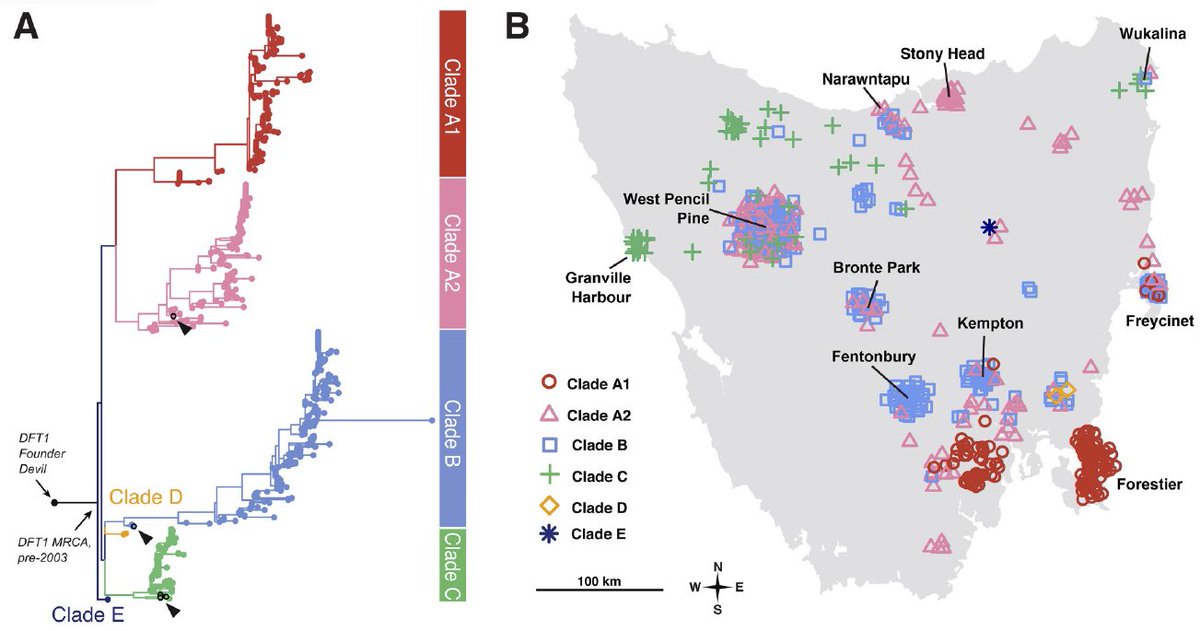 https://doi.org/10.1371/j..." title="Hi Science Twitter!A big shout out to my two fantastic colleagues Young Mi and Kevin, who have just released what is likely the largest tumour phylogeny built to date: the tree of a SINGLE cancer, recreated from >600 biopsies! A thread (1/10)https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Rückhand Zeigefinger nach unten" aria-label="Emoji: Rückhand Zeigefinger nach unten"> https://doi.org/10.1371/j..." class="img-responsive" style="max-width:100%;"/>
https://doi.org/10.1371/j..." title="Hi Science Twitter!A big shout out to my two fantastic colleagues Young Mi and Kevin, who have just released what is likely the largest tumour phylogeny built to date: the tree of a SINGLE cancer, recreated from >600 biopsies! A thread (1/10)https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Rückhand Zeigefinger nach unten" aria-label="Emoji: Rückhand Zeigefinger nach unten"> https://doi.org/10.1371/j..." class="img-responsive" style="max-width:100%;"/>








 https://abs.twimg.com/emoji/v2/... draggable="false" alt="🥾" title="Hiking boot" aria-label="Emoji: Hiking boot">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌏" title="Asien-Australien auf dem Globus" aria-label="Emoji: Asien-Australien auf dem Globus">(10/10)" title="Super happy to be part of this study over the last ~5 years! Thanks to our mentor Liz Murchison, wonderful colleagues and friends at UTAS/STDP/Menzies, Kevin, Young Mi and I could even join for a number of devil monitoring adventures in Tasmania ourselveshttps://abs.twimg.com/emoji/v2/... draggable="false" alt="🐾" title="Pfotenabdrücke" aria-label="Emoji: Pfotenabdrücke">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🥾" title="Hiking boot" aria-label="Emoji: Hiking boot">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌏" title="Asien-Australien auf dem Globus" aria-label="Emoji: Asien-Australien auf dem Globus">(10/10)">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🥾" title="Hiking boot" aria-label="Emoji: Hiking boot">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌏" title="Asien-Australien auf dem Globus" aria-label="Emoji: Asien-Australien auf dem Globus">(10/10)" title="Super happy to be part of this study over the last ~5 years! Thanks to our mentor Liz Murchison, wonderful colleagues and friends at UTAS/STDP/Menzies, Kevin, Young Mi and I could even join for a number of devil monitoring adventures in Tasmania ourselveshttps://abs.twimg.com/emoji/v2/... draggable="false" alt="🐾" title="Pfotenabdrücke" aria-label="Emoji: Pfotenabdrücke">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🥾" title="Hiking boot" aria-label="Emoji: Hiking boot">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌏" title="Asien-Australien auf dem Globus" aria-label="Emoji: Asien-Australien auf dem Globus">(10/10)">
 https://abs.twimg.com/emoji/v2/... draggable="false" alt="🥾" title="Hiking boot" aria-label="Emoji: Hiking boot">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌏" title="Asien-Australien auf dem Globus" aria-label="Emoji: Asien-Australien auf dem Globus">(10/10)" title="Super happy to be part of this study over the last ~5 years! Thanks to our mentor Liz Murchison, wonderful colleagues and friends at UTAS/STDP/Menzies, Kevin, Young Mi and I could even join for a number of devil monitoring adventures in Tasmania ourselveshttps://abs.twimg.com/emoji/v2/... draggable="false" alt="🐾" title="Pfotenabdrücke" aria-label="Emoji: Pfotenabdrücke">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🥾" title="Hiking boot" aria-label="Emoji: Hiking boot">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌏" title="Asien-Australien auf dem Globus" aria-label="Emoji: Asien-Australien auf dem Globus">(10/10)">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🥾" title="Hiking boot" aria-label="Emoji: Hiking boot">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌏" title="Asien-Australien auf dem Globus" aria-label="Emoji: Asien-Australien auf dem Globus">(10/10)" title="Super happy to be part of this study over the last ~5 years! Thanks to our mentor Liz Murchison, wonderful colleagues and friends at UTAS/STDP/Menzies, Kevin, Young Mi and I could even join for a number of devil monitoring adventures in Tasmania ourselveshttps://abs.twimg.com/emoji/v2/... draggable="false" alt="🐾" title="Pfotenabdrücke" aria-label="Emoji: Pfotenabdrücke">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🥾" title="Hiking boot" aria-label="Emoji: Hiking boot">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌏" title="Asien-Australien auf dem Globus" aria-label="Emoji: Asien-Australien auf dem Globus">(10/10)">
 https://abs.twimg.com/emoji/v2/... draggable="false" alt="🥾" title="Hiking boot" aria-label="Emoji: Hiking boot">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌏" title="Asien-Australien auf dem Globus" aria-label="Emoji: Asien-Australien auf dem Globus">(10/10)" title="Super happy to be part of this study over the last ~5 years! Thanks to our mentor Liz Murchison, wonderful colleagues and friends at UTAS/STDP/Menzies, Kevin, Young Mi and I could even join for a number of devil monitoring adventures in Tasmania ourselveshttps://abs.twimg.com/emoji/v2/... draggable="false" alt="🐾" title="Pfotenabdrücke" aria-label="Emoji: Pfotenabdrücke">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🥾" title="Hiking boot" aria-label="Emoji: Hiking boot">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌏" title="Asien-Australien auf dem Globus" aria-label="Emoji: Asien-Australien auf dem Globus">(10/10)">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🥾" title="Hiking boot" aria-label="Emoji: Hiking boot">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌏" title="Asien-Australien auf dem Globus" aria-label="Emoji: Asien-Australien auf dem Globus">(10/10)" title="Super happy to be part of this study over the last ~5 years! Thanks to our mentor Liz Murchison, wonderful colleagues and friends at UTAS/STDP/Menzies, Kevin, Young Mi and I could even join for a number of devil monitoring adventures in Tasmania ourselveshttps://abs.twimg.com/emoji/v2/... draggable="false" alt="🐾" title="Pfotenabdrücke" aria-label="Emoji: Pfotenabdrücke">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🥾" title="Hiking boot" aria-label="Emoji: Hiking boot">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌏" title="Asien-Australien auf dem Globus" aria-label="Emoji: Asien-Australien auf dem Globus">(10/10)">


