Would you want to screen a population of asymptomatic people with qPCR primers that have 6base pair 3 primer dimers?
The WHO recommends these primers.
@MichaelYeadon3 @ClareCraigPath
The WHO recommends these primers.
@MichaelYeadon3 @ClareCraigPath
I’m not going to say this is causing False positives or False negatives as that requires wet lab work to really know but primer dimer screens is PCR 101 and it’s not in place for primers from the WHO.
The other PCR 101 screen is BLAST against an off target database.
The other PCR 101 screen is BLAST against an off target database.
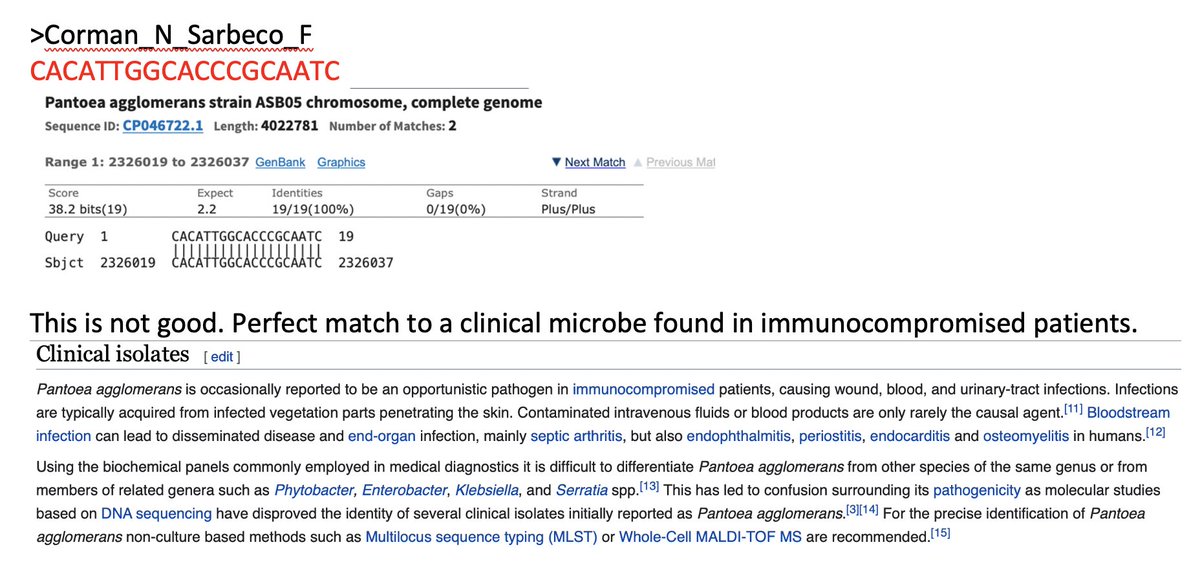
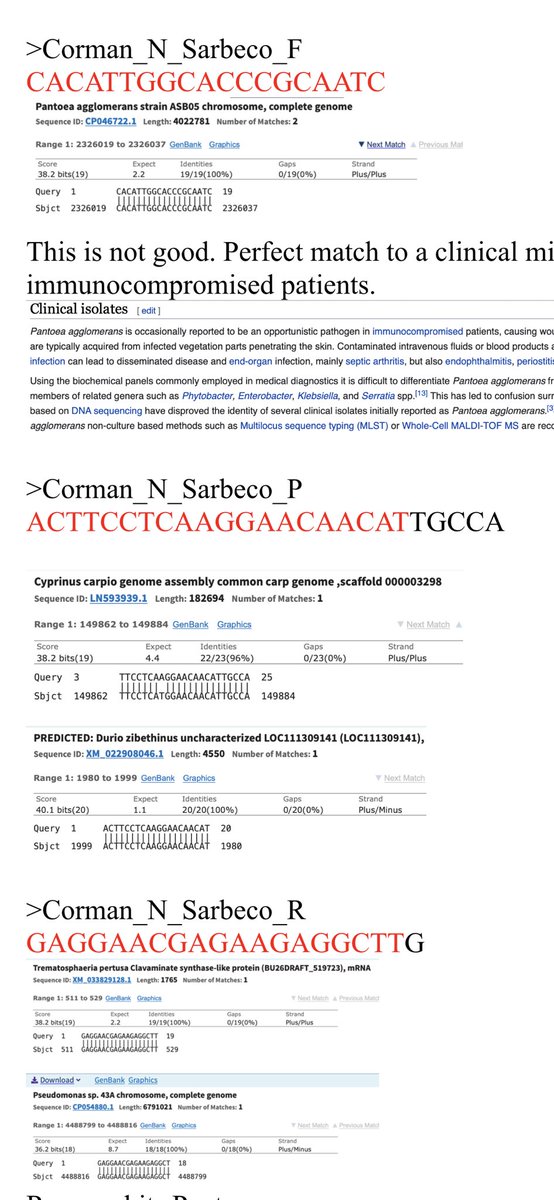
Having 100% match of a primer to Pantoea is a yellow flag.
It’s 1/2 of a red flag. You need both primers and probes to land in proximity for amplification and the reverse primer lands in Pantoea far away....
But we’re doing RT-PCR. Distant parts of the genome can be close in RNA.
It’s 1/2 of a red flag. You need both primers and probes to land in proximity for amplification and the reverse primer lands in Pantoea far away....
But we’re doing RT-PCR. Distant parts of the genome can be close in RNA.
If you want to screen for primer Dimers.
Plug the primer sequences into this Thermo web tool.
Thermo did $2B in COVID Testing last quarter. They understand PCR. https://www.thermofisher.com/us/en/home/brands/thermo-scientific/molecular-biology/molecular-biology-learning-center/molecular-biology-resource-library/thermo-scientific-web-tools/multiple-primer-analyzer.html">https://www.thermofisher.com/us/en/hom...
Plug the primer sequences into this Thermo web tool.
Thermo did $2B in COVID Testing last quarter. They understand PCR. https://www.thermofisher.com/us/en/home/brands/thermo-scientific/molecular-biology/molecular-biology-learning-center/molecular-biology-resource-library/thermo-scientific-web-tools/multiple-primer-analyzer.html">https://www.thermofisher.com/us/en/hom...
>Corman_RdRp_SARs_F1
GTGAAATGGTCATGTGTGGCGG
>Corman_RdRp_SARs_F2
GTGAGATGGTCATGTGTGGCGG
>Corman_RdRp_SARSr-P2.a
CCAGGTGGAACATCATCAGGTGATGC
>Corman_RdRp_SARSr-P2.a1
CCAGGTGGAACGTCATCAGGTGATGC
>Corman_RdRp_SARSr-P2.b
CCAGGTGGAACATCATCCGGTGATGC
GTGAAATGGTCATGTGTGGCGG
>Corman_RdRp_SARs_F2
GTGAGATGGTCATGTGTGGCGG
>Corman_RdRp_SARSr-P2.a
CCAGGTGGAACATCATCAGGTGATGC
>Corman_RdRp_SARSr-P2.a1
CCAGGTGGAACGTCATCAGGTGATGC
>Corman_RdRp_SARSr-P2.b
CCAGGTGGAACATCATCCGGTGATGC
>Corman_RdRp_SARSr-P2.b1
CCAGGTGGAACGTCATCCGGTGATGC
>Corman_RdRp_SARSr-P2.c
CCAGGTGGTACATCATCAGGTGATGC
>Corman_RdRp_SARSr-P2.c1
CCAGGTGGTACGTCATCAGGTGATGC
>Corman_RdRp_SARSr-P2.d
CCAGGTGGTACATCATCCGGTGATGC
>Corman_RdRp_SARSr-P2.d
CCAGGTGGTACGTCATCCGGTGATGC
CCAGGTGGAACGTCATCCGGTGATGC
>Corman_RdRp_SARSr-P2.c
CCAGGTGGTACATCATCAGGTGATGC
>Corman_RdRp_SARSr-P2.c1
CCAGGTGGTACGTCATCAGGTGATGC
>Corman_RdRp_SARSr-P2.d
CCAGGTGGTACATCATCCGGTGATGC
>Corman_RdRp_SARSr-P2.d
CCAGGTGGTACGTCATCCGGTGATGC
>Corman_RdRp_SARSr-R
CAAATGTTAAACACACTATTAGCATA
>Corman_RdRp_SARSr-R2
CAAATGTTAAACACACTATTAGCATA
>Corman_RdRp_SARSr-R3
CAGATGTTAAAGACACTATTAGCATA
>Corman_RdRp_SARSr-R4
CAGATGTTAAAGACACTATTAGCATA
>Corman_E_Sarbeco_F
ACAGGTACGTTAATAGTTAATAGCGT
CAAATGTTAAACACACTATTAGCATA
>Corman_RdRp_SARSr-R2
CAAATGTTAAACACACTATTAGCATA
>Corman_RdRp_SARSr-R3
CAGATGTTAAAGACACTATTAGCATA
>Corman_RdRp_SARSr-R4
CAGATGTTAAAGACACTATTAGCATA
>Corman_E_Sarbeco_F
ACAGGTACGTTAATAGTTAATAGCGT
>Corman_E_Sarbeco_P1
ACACTAGCCATCCTTACTGCGCTTCG
>Corman_E_Sarbeco_R
ATATTGCAGCAGTACGCACACA
>Corman_N_Sarbeco_F
CACATTGGCACCCGCAATC
>Corman_N_Sarbeco_P
ACTTCCTCAAGGAACAACATTGCCA
>Corman_N_Sarbeco_R
GAGGAACGAGAAGAGGCTTG
ACACTAGCCATCCTTACTGCGCTTCG
>Corman_E_Sarbeco_R
ATATTGCAGCAGTACGCACACA
>Corman_N_Sarbeco_F
CACATTGGCACCCGCAATC
>Corman_N_Sarbeco_P
ACTTCCTCAAGGAACAACATTGCCA
>Corman_N_Sarbeco_R
GAGGAACGAGAAGAGGCTTG
That’s right. There are 20 primers in the mix because the protocol has degenerate bases in the primers (R,S,W,M).
This increases primer dimer potential and off target PCR.
That alone isn’t lethal but jack the primer conc, the dNTPs, the Mg, use a low anneal temp...
This increases primer dimer potential and off target PCR.
That alone isn’t lethal but jack the primer conc, the dNTPs, the Mg, use a low anneal temp...
While having primers with wide annealing temps and you better run a primer dimer and BLAST screen.
Pandemic timelines cut corners. I can understand that. But I can’t seem to figure what lab is running what test 10 months later and the FP discussions go in circles.
Pandemic timelines cut corners. I can understand that. But I can’t seem to figure what lab is running what test 10 months later and the FP discussions go in circles.
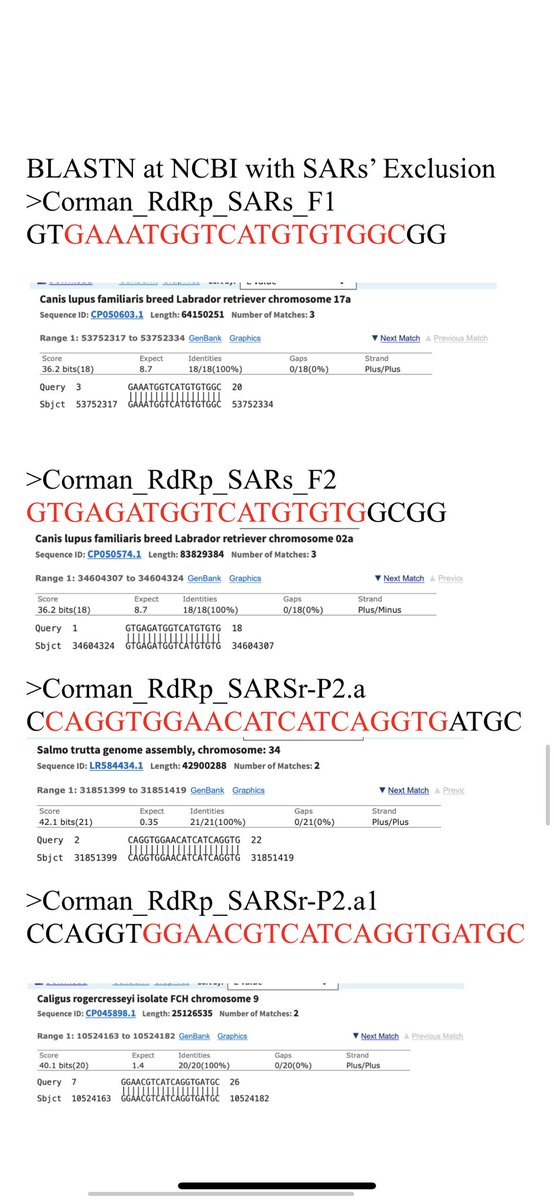
Here are a few more homologies of the primers to off target organisms.
There are some long stretches but unless the 3’ end of the primer is landing down these may only recruit primer out of solution and lower efficiency. Might explain the high primer conc used.
There are some long stretches but unless the 3’ end of the primer is landing down these may only recruit primer out of solution and lower efficiency. Might explain the high primer conc used.
In some cases the 3’ end lands. This is only a problem if the organism is believed to be a potential background organism. We don’t expect Brown Trout to be in your nose. Pantoea could be there.
There is a long list of hits. I’ve only highlighted a few.
You can copy and paste the above primers into NCBI BLAST and have a hay day poking through potential hits.
https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome">https://blast.ncbi.nlm.nih.gov/Blast.cgi...
You can copy and paste the above primers into NCBI BLAST and have a hay day poking through potential hits.
https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome">https://blast.ncbi.nlm.nih.gov/Blast.cgi...
Plug the sequences into the top box.
Select the exclude button and select SARs.
Leave it on default NR as a target database. There are other that might expand the search but this is a good place to start.
Radio button at the bottom on blastn.
Hit BLAST and wait.
Select the exclude button and select SARs.
Leave it on default NR as a target database. There are other that might expand the search but this is a good place to start.
Radio button at the bottom on blastn.
Hit BLAST and wait.
You’ll get a long list of hits. Might explain some of the examples of people sending in test kits with Papaya and getting positive results?
But I really can’t confirm that as its not clear to me which labs are running which tests at what CQ thresholds.
But I really can’t confirm that as its not clear to me which labs are running which tests at what CQ thresholds.
The WHO protocol does have an update removing the N gene.
But thats not the primer causing the dimers.
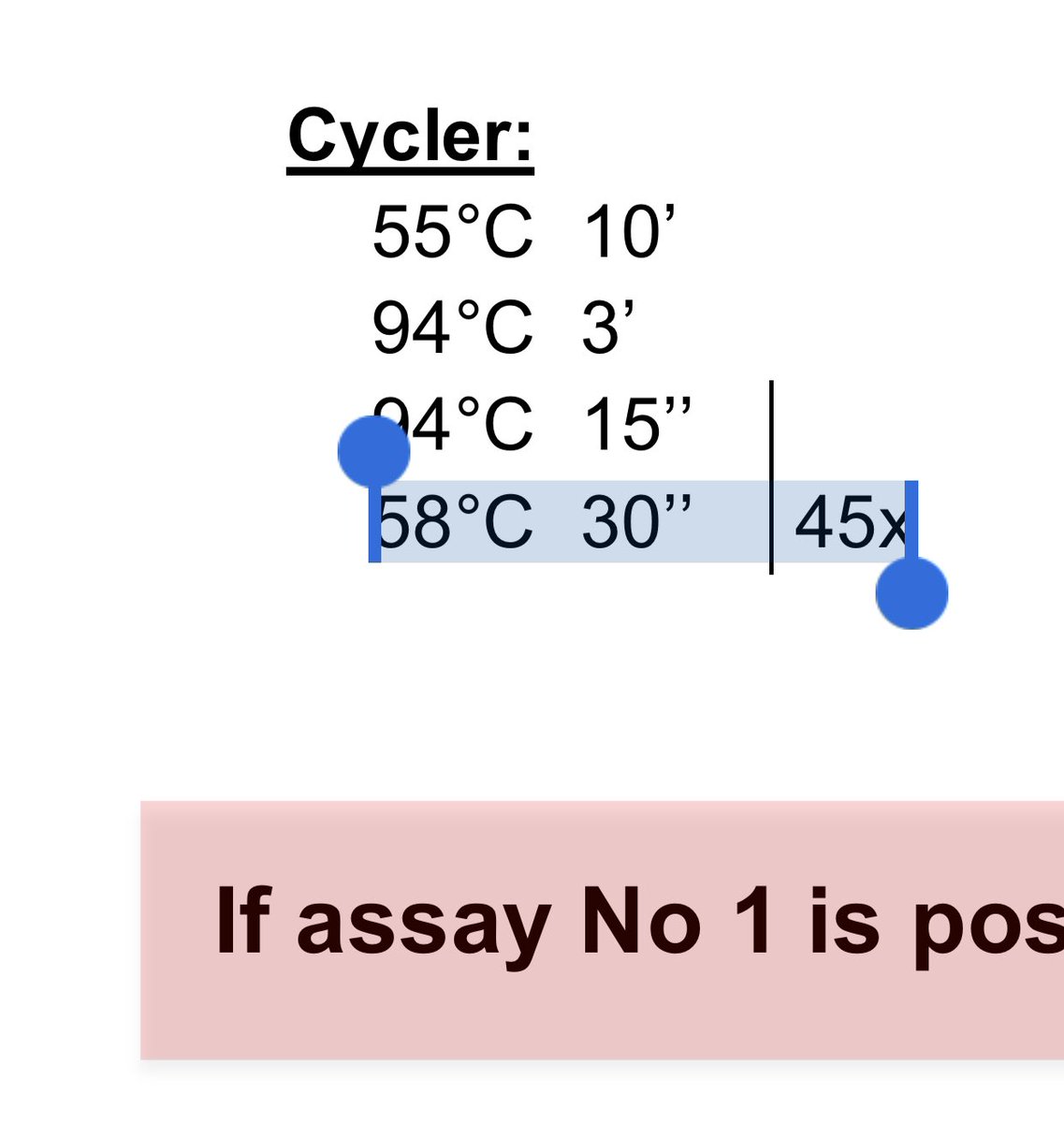
58C annealing is cold and sticky. Spike in some Mg & even stickier.
45 cycles of it with zero recommended CQ threshold is asking for mass confusion.
https://www.who.int/docs/default-source/coronaviruse/protocol-v2-1.pdf">https://www.who.int/docs/defa...
But thats not the primer causing the dimers.
58C annealing is cold and sticky. Spike in some Mg & even stickier.
45 cycles of it with zero recommended CQ threshold is asking for mass confusion.
https://www.who.int/docs/default-source/coronaviruse/protocol-v2-1.pdf">https://www.who.int/docs/defa...
When I’m really down and my PCR isn’t working...
I would run a gradient PCR 58-68C to try to find temps that amplify the target.
Bump the cycles to 45.
Add MgCl2
2x the nucleotides and the primers.
Pretty much whats seen in here.
But a day In BLAST is worth a week in the lab.
I would run a gradient PCR 58-68C to try to find temps that amplify the target.
Bump the cycles to 45.
Add MgCl2
2x the nucleotides and the primers.
Pretty much whats seen in here.
But a day In BLAST is worth a week in the lab.
Now Pantoea might be a stretch for your nose. It has some clinical lit but is usually sourced from a plant.
We see it in inhaled cannabis flower.
More relevant might be the streptomycese hit but it’s not a perfect 3’ hit.
This kit is recommended 45 cycles.
We see it in inhaled cannabis flower.
More relevant might be the streptomycese hit but it’s not a perfect 3’ hit.
This kit is recommended 45 cycles.
That doesn’t mean labs are calling CQ 45 positives. That’s a major straw man circulating right now.
But it’s a very valid point of confusion when protocols like this don’t specify the CQ cutoff and the WHO rubber stamps them.
If the WHO cant perform this type of analysis, STFU.
But it’s a very valid point of confusion when protocols like this don’t specify the CQ cutoff and the WHO rubber stamps them.
If the WHO cant perform this type of analysis, STFU.
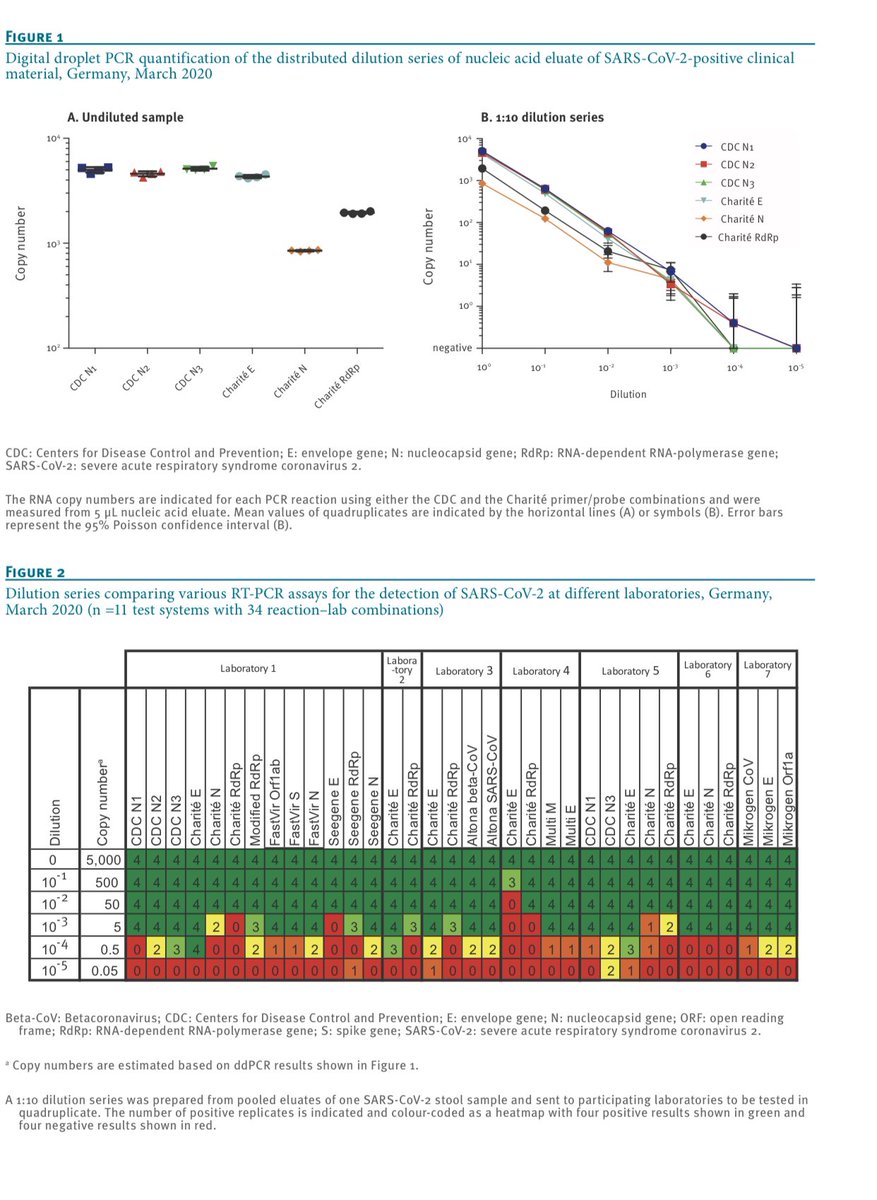
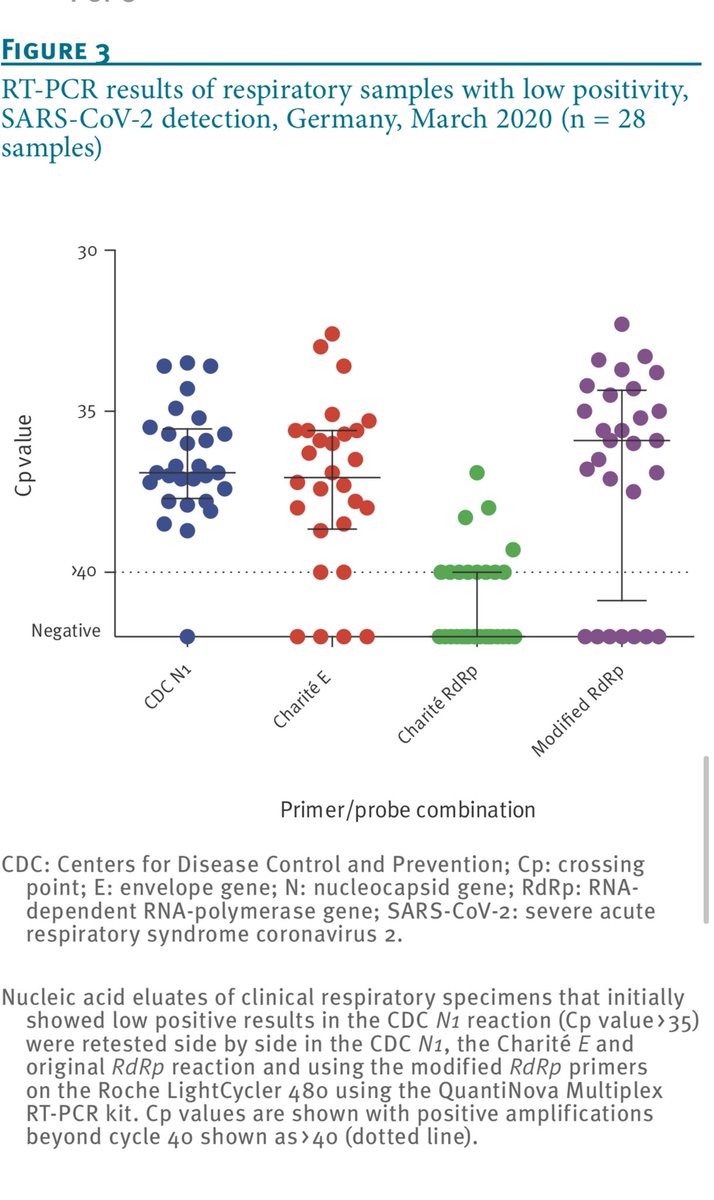
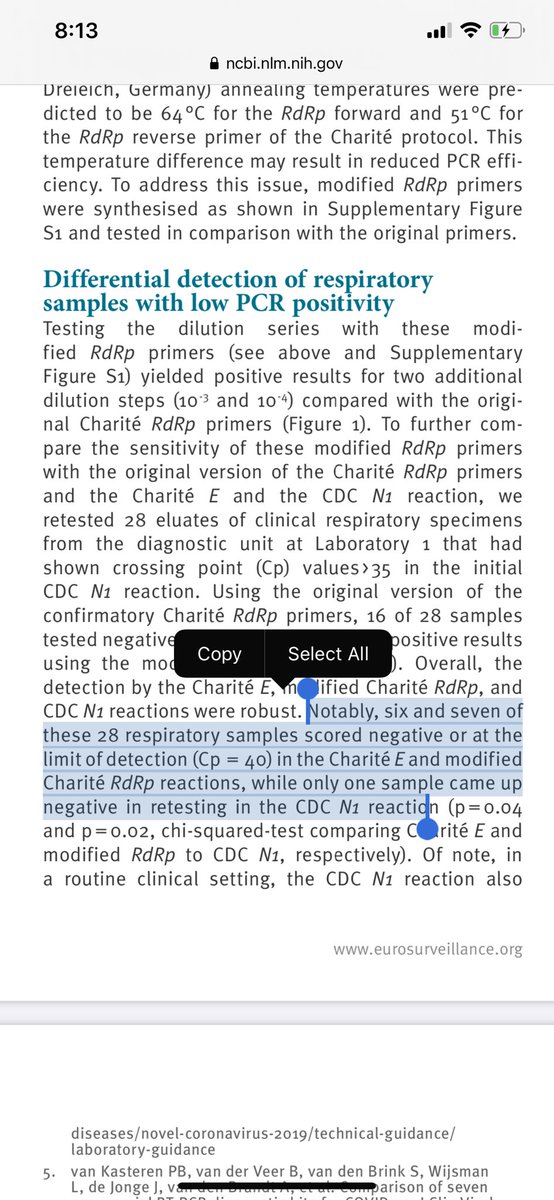
You can see these flaws manifest themselves in various ways in this comparison paper.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7315722/pdf/eurosurv-25-24-1.pdf">https://www.ncbi.nlm.nih.gov/pmc/artic...
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7315722/pdf/eurosurv-25-24-1.pdf">https://www.ncbi.nlm.nih.gov/pmc/artic...
They seem to manifest in lower detection sensitivity.
Many may confuse this with less false positives but that it isn’t always the case.
You can have low efficiency on your target because of off target efficiency (primer dimers etc).
Does the off target PCR create signal?
Many may confuse this with less false positives but that it isn’t always the case.
You can have low efficiency on your target because of off target efficiency (primer dimers etc).
Does the off target PCR create signal?
It is possible to have a kit under detect true positives and still have false positives that are triggered by bad primer design
This would be easier to know if labs were more transparent on the kits they use and CQ they call at.
Otherwise people are left to falsely assume 45.
This would be easier to know if labs were more transparent on the kits they use and CQ they call at.
Otherwise people are left to falsely assume 45.
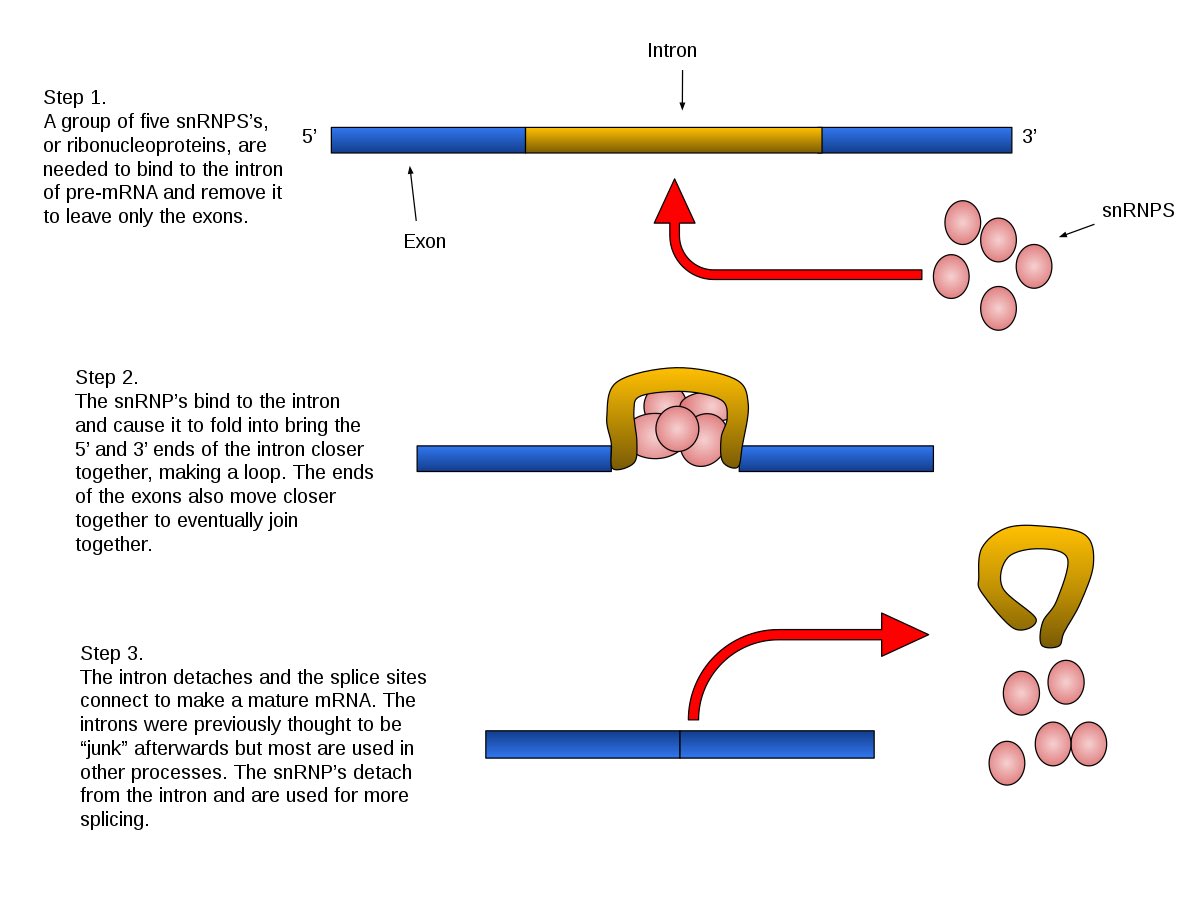
This is an important point when designing RTqPCR kits as the primers can amplify both the transcriptome & genomes of the background sample. Transcriptomes of eukaryotes often splice distant regions of the genome together. Notice the hits to canine and rhesus. Fungi come to mind.
What is the point of an FDA EUA approval process if consumers can’t figure out what test their lab is running?
This sounds like a big game of “Trust us. The FDA looked at this. There is nothing to see here.”
I can easily find EUA docs at the FDA site and this is about the WHO.
This sounds like a big game of “Trust us. The FDA looked at this. There is nothing to see here.”
I can easily find EUA docs at the FDA site and this is about the WHO.
So I’m not blaming the FDA for these WHO primers but I don’t see the point in the EUA approval process if people versed in this field can’t figure out what test is being run on their children to quarantine them.
When transparency lacks, civility becomes tyranny.
When transparency lacks, civility becomes tyranny.

 Read on Twitter
Read on Twitter