We are excited to share the @bbglab contribution to "Mutational dynamics and transmission properties of SARS-CoV-2". An interdisciplinary collaborative effort studying the mutational dynamics of #SARSCoV2 in Austria. @CeMM_News @Bergthalerlab
https://stm.sciencemag.org/content/early/2020/11/20/scitranslmed.abe2555/">https://stm.sciencemag.org/content/e... https://abs.twimg.com/emoji/v2/... draggable="false" alt="🧵" title="Thread" aria-label="Emoji: Thread">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🧵" title="Thread" aria-label="Emoji: Thread"> https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Rückhand Zeigefinger nach unten" aria-label="Emoji: Rückhand Zeigefinger nach unten">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Rückhand Zeigefinger nach unten" aria-label="Emoji: Rückhand Zeigefinger nach unten">
https://stm.sciencemag.org/content/early/2020/11/20/scitranslmed.abe2555/">https://stm.sciencemag.org/content/e...
A compendium of different analyses, combining epidemiology with deep viral genome sequencing points to mutational dynamics, transmission properties, and reconstruction of superspreading events. The paper is very interesting!
In this thread I will only focus on our contribution https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Rückhand Zeigefinger nach unten" aria-label="Emoji: Rückhand Zeigefinger nach unten">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Rückhand Zeigefinger nach unten" aria-label="Emoji: Rückhand Zeigefinger nach unten">
In this thread I will only focus on our contribution
Given the exceptional circumstances and our curiosity in March we started analysing SARS-CoV-2 genomes using our @bbglab expertise on cancer genomics to learn how mutations accumulate in the virus genome. We joined the collaborative effort led by @Bergthalerlab at @CeMM_News
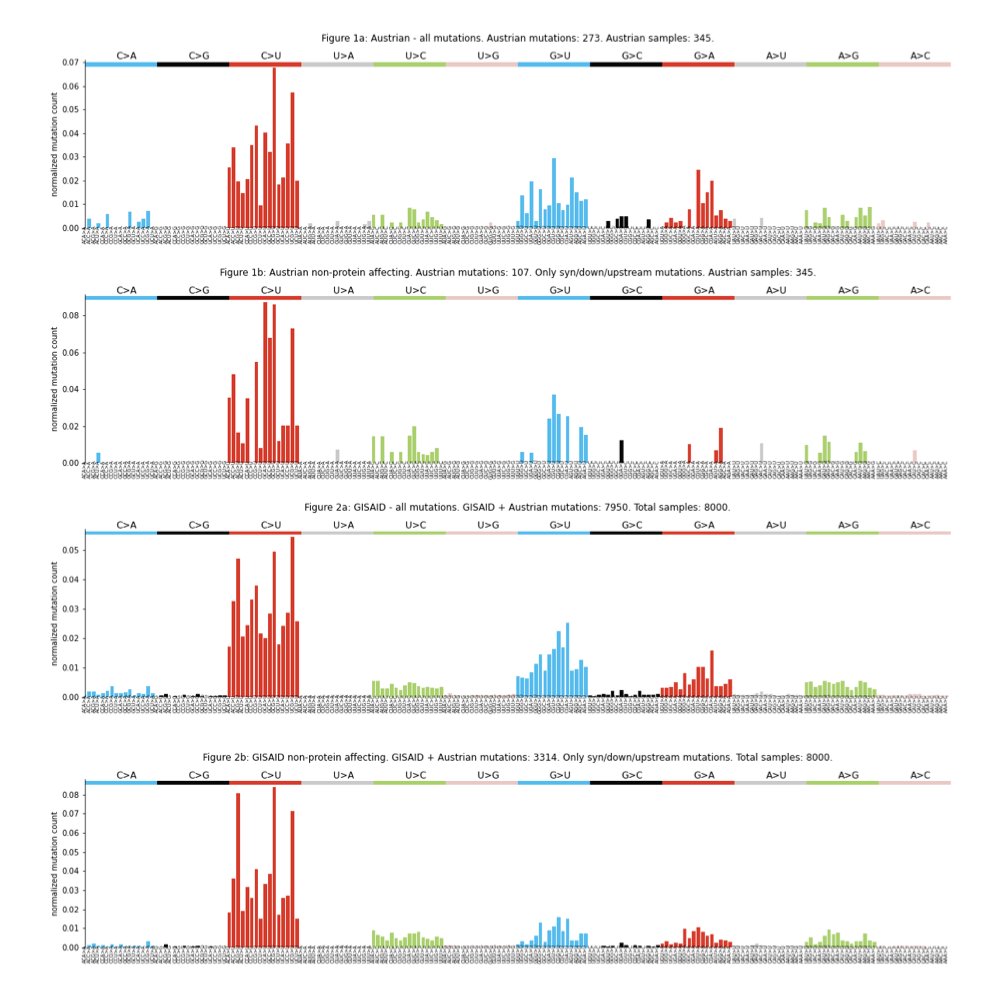
We first looked into the mutational profile of SARS-CoV-2 variants and observed a strong bias towards C>U, G>A, G>U mutations, with some U>C and A>G. This is maintained in synonymous variants, suggesting it is not a result of selection but of the operating mutational processes
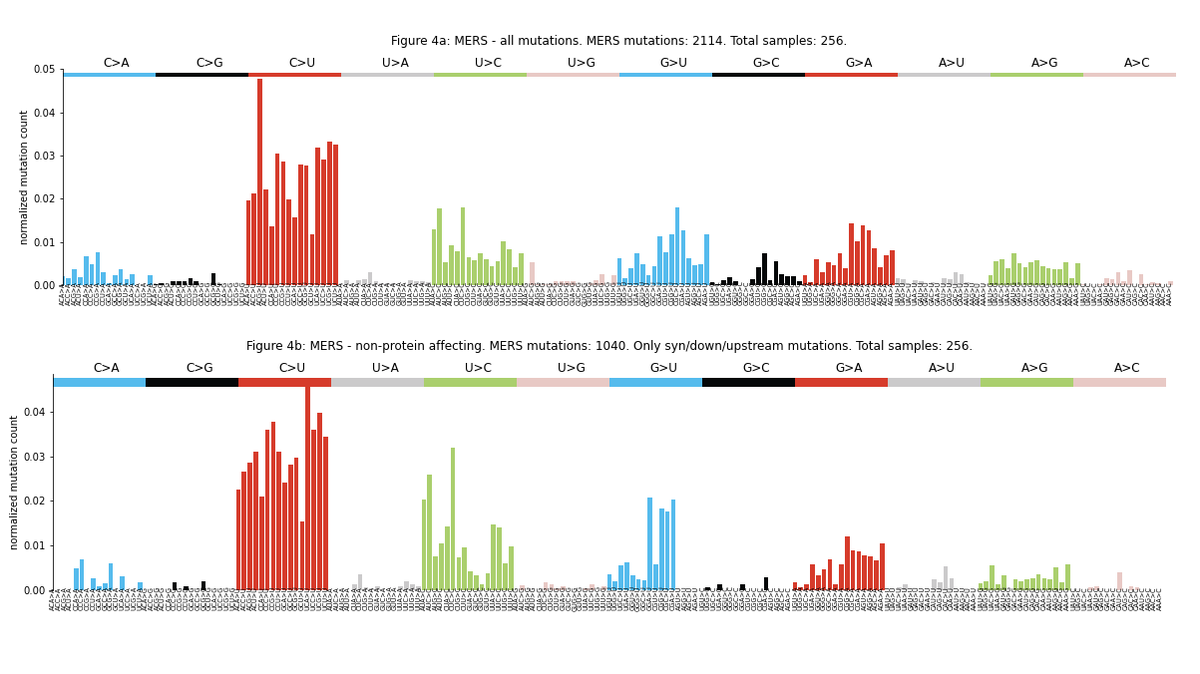
The mutational profile is consistent across different sequencing platforms. Interestingly, it is also highly similar to the MERS profile, suggesting that similar mutational processes may be operating in both viruses.
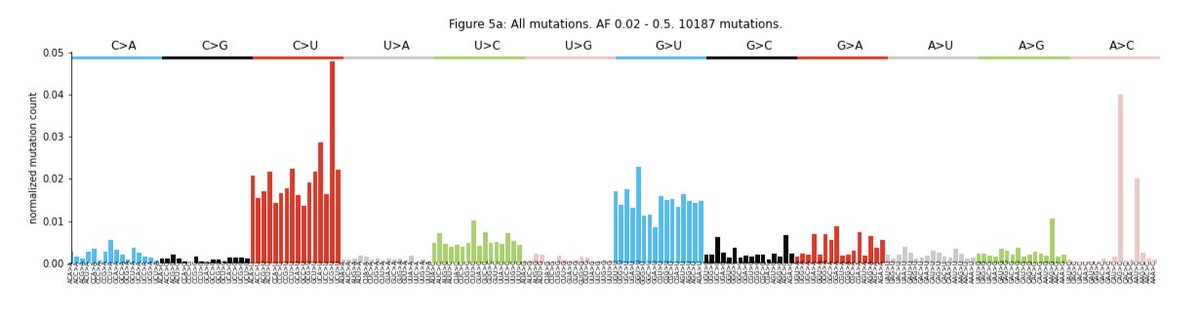
Next we asked if this mutational profile was consistent across variants within the hosts (intra-host), and it is! We probed different allele frequencies (AF) and a similar profile is observed within the 0.02 - 0.5 AF range.
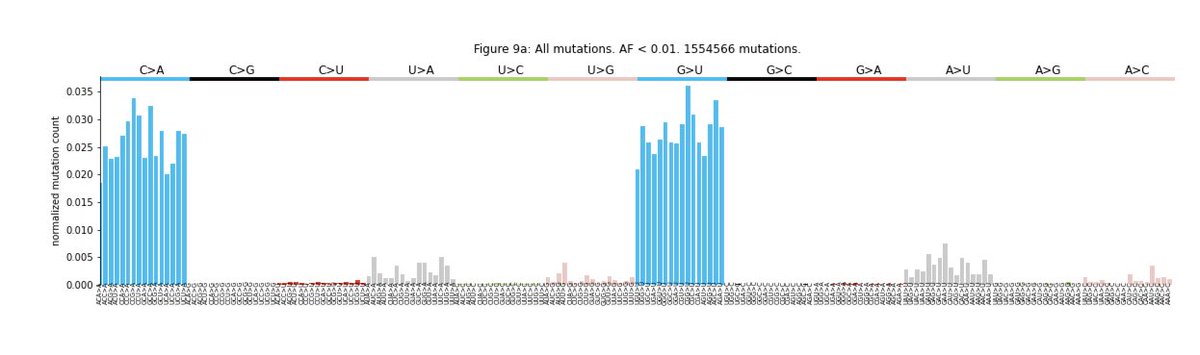
In contrast intra-host variants with AF<0.02 show strong bias in C>A and G>U transversions, indicating possible artifactual mutations. For this reason the rest of the analyses in the manuscript were done with mutations with AF > 0.02.
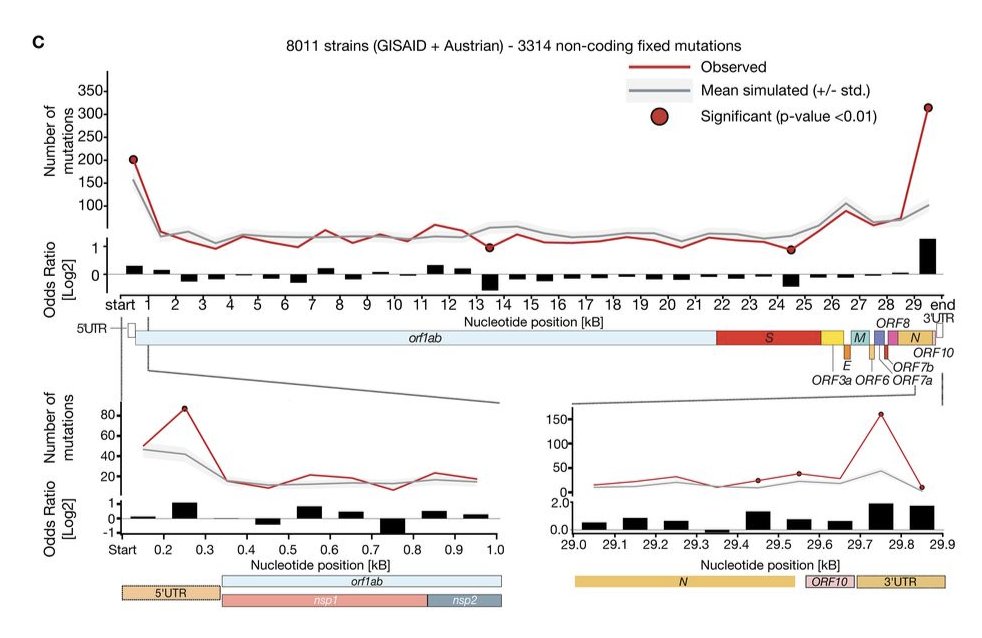
We also wondered whether the mutation rate across the viral genome was homogeneous. According to the data, it seems that UTRs have a higher mutation burden, which in part (but not entirely) is explained by the tri-nucleotide composition.
When we did the analysis for the paper there wasn’t enough data to do mutational signature extraction. But as SARS-CoV-2 data accumulates quickly we now have enough mutations from @CovidGenomicsUK to perform mutational signature extraction. Results in: https://github.com/bbglab/sars-cov-2-genomics/">https://github.com/bbglab/sa...
One extracted signature is highly similar to the SBS2 caused by APOBEC, another is dominated by A>G changes, which could be caused by the ADAR family of deaminases. This agrees with previous reports that proposed RNA editing enzymes as main mutagenic process in SARS-CoV-2
All our code, data and results are available at https://zenodo.org/record/4275398 ">https://zenodo.org/record/42... and https://github.com/bbglab/sars-cov-2-genomics/">https://github.com/bbglab/sa... We are open to comments, ideas and suggestions.
More information about the consortium: https://www.sarscov2-austria.org/ ">https://www.sarscov2-austria.org/">...
More information about the consortium: https://www.sarscov2-austria.org/ ">https://www.sarscov2-austria.org/">...
We feel fortunate to have had the opportunity to contribute to this work with a great interdisciplinary team of scientists from @Bergthalerlab, @Alex_M_Popa, @DerGenger, @abergthaler, @Harvard, @MedUni_Wien, @DanaFarber, @univienna, Kaiser Franz Josef Hospital, @agesnews.
At @bbglab this work has been a collaborative effort between @fran_mj88, @oriol_pich, @nlbigas and myself

 Read on Twitter
Read on Twitter " title="A compendium of different analyses, combining epidemiology with deep viral genome sequencing points to mutational dynamics, transmission properties, and reconstruction of superspreading events. The paper is very interesting!In this thread I will only focus on our contribution https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Rückhand Zeigefinger nach unten" aria-label="Emoji: Rückhand Zeigefinger nach unten">" class="img-responsive" style="max-width:100%;"/>
" title="A compendium of different analyses, combining epidemiology with deep viral genome sequencing points to mutational dynamics, transmission properties, and reconstruction of superspreading events. The paper is very interesting!In this thread I will only focus on our contribution https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Rückhand Zeigefinger nach unten" aria-label="Emoji: Rückhand Zeigefinger nach unten">" class="img-responsive" style="max-width:100%;"/>