Recently reported #SARSCoV2 #COVID19 & #39; #mink mutation& #39; in #Denmark is getting a lot of attention right now, but officially we know almost nothing about the variants/mutations in question.
From an SSI report from Sept, here is a little info.
https://www.ssi.dk/-/media/arkiv/dk/aktuelt/sygdomsudbrud/covid19/besvarelse-virkemidler-til-at-forebygge-smitte-mink-menneske.pdf?la=da
1/21">https://www.ssi.dk/-/media/a...
From an SSI report from Sept, here is a little info.
https://www.ssi.dk/-/media/arkiv/dk/aktuelt/sygdomsudbrud/covid19/besvarelse-virkemidler-til-at-forebygge-smitte-mink-menneske.pdf?la=da
1/21">https://www.ssi.dk/-/media/a...
*Very important caveat* here that I& #39;m working with very limited information, but trying to make some sense of something that& #39;s getting a lot of attention, with the little info I can find. However, this should be taken as a tweet thread, not a preprint or anything similar.
2/21
2/21
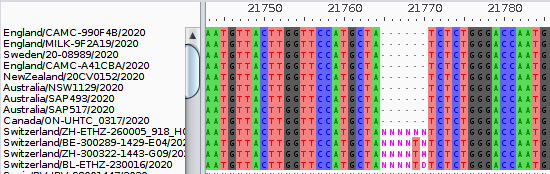
On page 8, the document describes 2 variants, defined by mutations A22920T & a 2 amino-acid deletion from 21766-21771. Both are changes in the Spike protein.
3/21
3/21
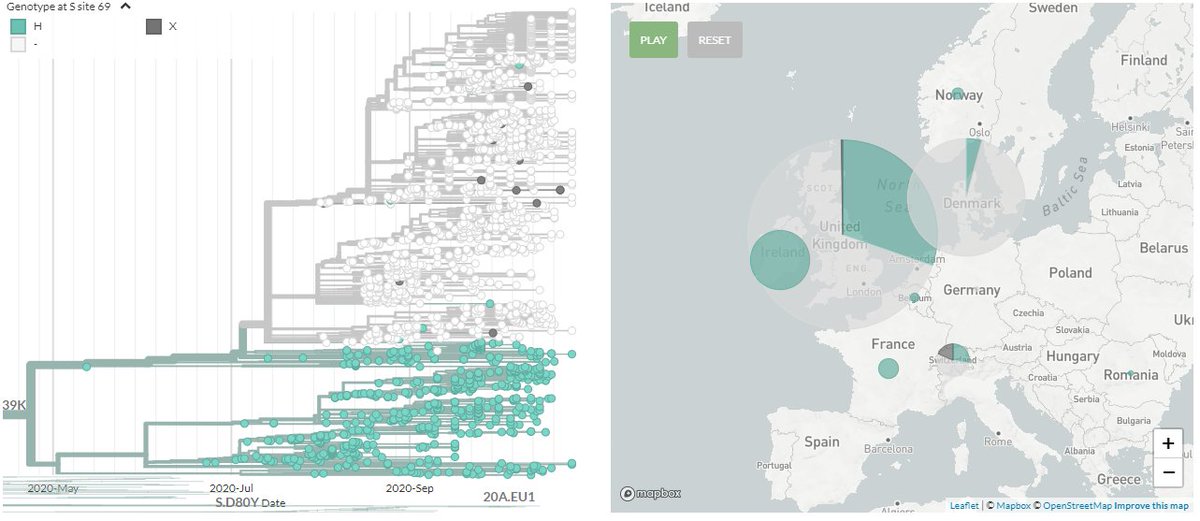
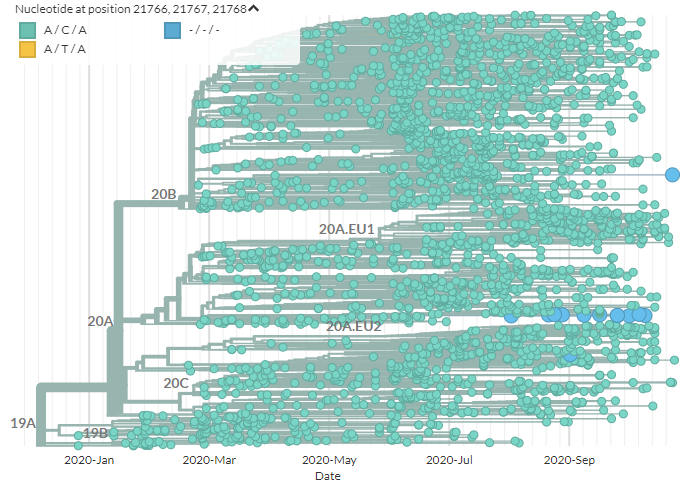
First, we& #39;ll look at the deletion. In the Neher Lab Europe build, we can color by the first 3 positions to identify sequences with the deletion (blue).
We can see it a few places, but one distinct cluster, at the centre.
4/21
https://nextstrain.org/groups/neherlab/ncov/europe?c=gt-nuc_21766,21767,21768&gmax=22722&gmin=20823">https://nextstrain.org/groups/ne...
We can see it a few places, but one distinct cluster, at the centre.
4/21
https://nextstrain.org/groups/neherlab/ncov/europe?c=gt-nuc_21766,21767,21768&gmax=22722&gmin=20823">https://nextstrain.org/groups/ne...
If we zoom in on this cluster, we can see it& #39;s found across Europe & at least once in Australia, & that it has the deletion.
5/21
5/21
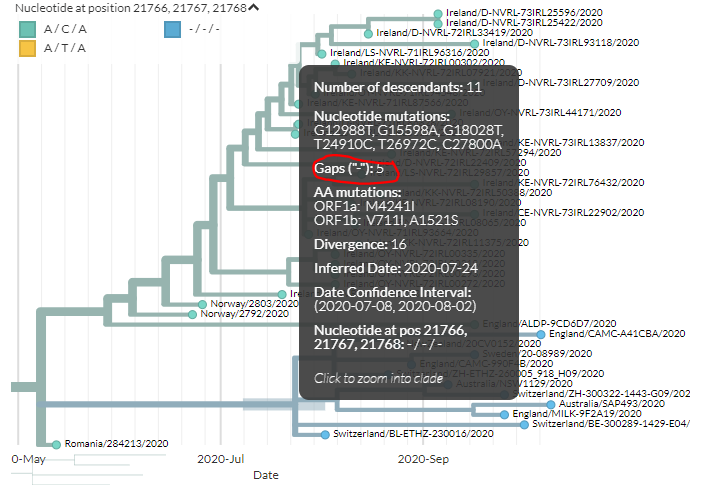
As a bit of a side note - I don& #39;t colour by all positions because in some sequences the deletions seem to have been at least partially filled - the colours on the tree get more complicated. We& #39;ve just now confirmed with the sequencers this is really a full deletion.
6/21
6/21
This blue & #39;deletion& #39; cluster is part of a larger cluster - one with the N439K mutation in Spike - one that already has caught the interest of scientists.
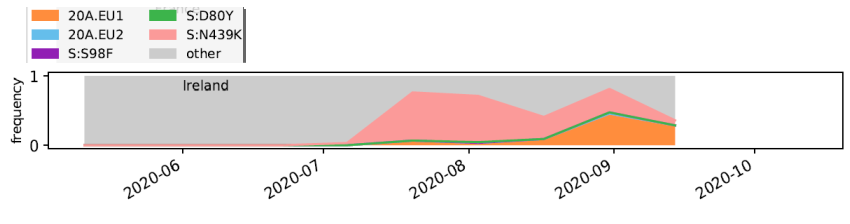
This cluster with N439K is found across Europe, including Ireland, where it has been fairly prevalent (pink).
7/21
This cluster with N439K is found across Europe, including Ireland, where it has been fairly prevalent (pink).
7/21
In my own build of N439K (sorry, not online yet), which contains every #SARSCoV2 sequence with this mutation, I can also color by these 3 deletion sites (yellow). About half of the N439K mutations contain this deletion.
8/21
8/21
We can see this deletion in Australia, the UK, Switzerland, Germany, & Sweden. It could be in more places, but we may lack sequences.
9/21
9/21
Back to N439K more generally. Scientists are already paying close attention to this mutation. It sits on the very top of Spike (a very & #39;sticky-out& #39; bit). It is part of the epitope for some neutralizing antibodies.
10/21
10/21
N439K has been reported to confer partial resistance to some antibodies.
SSI may have found similar outcomes in their research, if the variant they characterise by the deletions carries N439K, as almost all those we see do.
https://elifesciences.org/articles/61312
https://elifesciences.org/articles/... href=" https://www.nature.com/articles/s41586-020-2852-1
11/21">https://www.nature.com/articles/...
SSI may have found similar outcomes in their research, if the variant they characterise by the deletions carries N439K, as almost all those we see do.
https://elifesciences.org/articles/61312
11/21">https://www.nature.com/articles/...
Very important to highlight here that N439K conferring some level of resistance to an antibody is *not* necessarily the same as influencing the efficacy of a vaccine. The two should *not* be conflated.
12/21
12/21
It could also be that the deletion itself may have an effect - I& #39;m not aware of work on this yet. The deletion would be near the start of the Spike protein - my expertise is not structural, so I will not speculate further.
13/21
13/21
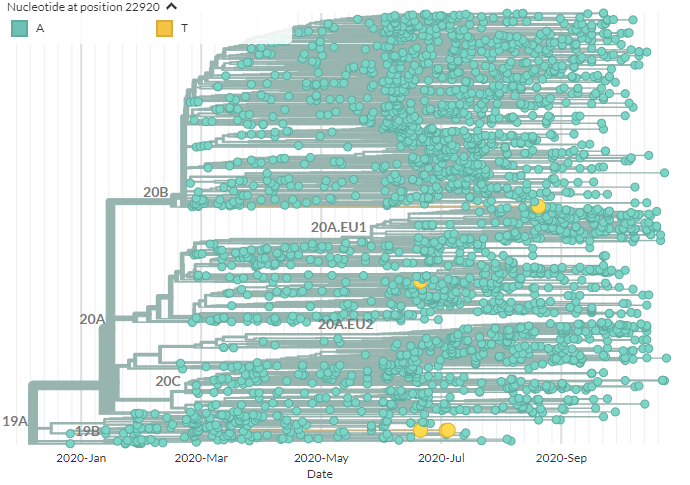
Now let& #39;s look at the A22920T mutation. This is Spike mutation Y453F.
We can color by this mutation on the same European tree (yellow). One sequence is from Russia, from a human. The others (middle & bottom)? From the Netherlands, in mink.
https://nextstrain.org/groups/neherlab/ncov/europe?c=gt-nuc_22920&gmax=22722&gmin=20823
14/21">https://nextstrain.org/groups/ne...
We can color by this mutation on the same European tree (yellow). One sequence is from Russia, from a human. The others (middle & bottom)? From the Netherlands, in mink.
https://nextstrain.org/groups/neherlab/ncov/europe?c=gt-nuc_22920&gmax=22722&gmin=20823
14/21">https://nextstrain.org/groups/ne...
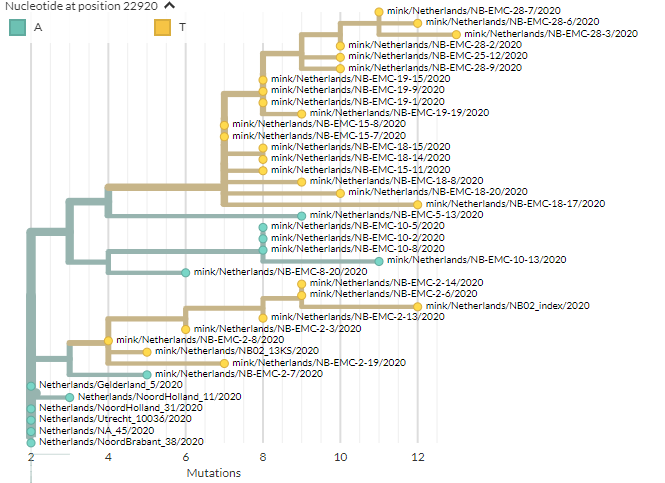
We can go to the Netherlands build to take a closer look. Again, 22920T is yellow.
https://nextstrain.org/groups/neherlab/ncov/netherlands?c=gt-nuc_22920
They& #39;re">https://nextstrain.org/groups/ne... all from mink. Not all mink have it, but we only see it in mink. (Zoomed view of bottom cluster to right)
15/21
https://nextstrain.org/groups/neherlab/ncov/netherlands?c=gt-nuc_22920
They& #39;re">https://nextstrain.org/groups/ne... all from mink. Not all mink have it, but we only see it in mink. (Zoomed view of bottom cluster to right)
15/21
It seems possible that the A22920T mutation may be a mink adaptation. It seems to have arisen multiple times in the mink samples we have from the Netherlands. Possibly it has also arisen in Denmark.
16/21
16/21
While we may find the same mutation in humans who& #39;ve been infected from a variant circulating or originating in mink, it might not have any impact. Indeed, it could be maladaptive (not work as well in humans if it& #39;s adapted to mink). We do not know either way yet.
17/21
17/21
It& #39;s import to repeat again that the above tweets are *my hypotheses* based on a Sept report from SSI which identifies 2 variants. More recent communications suggest there may be more mutations/variants detected now. Full information on what they have found is needed.
18/21
18/21
If Denmark believes this is serious enough to kill their entire mink population, one would perhaps also conclude that this serious enough to pass on the information about these mutations to scientists worldwide as quickly as possible to see if variants are found elsewhere.
19/21
19/21
It& #39;s critically important that novel #SARSCoV2 findings, particularly about spike mutations, are communicated responsibly. Announcements without context spark alarm, worry, & panic headlines. That doesn& #39;t help anyone, & scientists can& #39;t provide assessment or interpretation
20/21
20/21
Thanks to @richardneher for detective work assistance, @veeslerlab for info on 439, & Ivan Topolsky at ETH Zurich for confirming the deletion in Swiss sequences. And finally, to a kind follower for linking me to this very useful report.
21/21
21/21
A slightly more recent SSI paper from 2 Oct identifies the same two variants as the Sept report: A22920T/Y453F (mutation also found in NL mink) & deletion 21766-21771
https://www.ddd.dk/media/5662/dvk_epidemiologisk-udredning_ny.pdf
22/21">https://www.ddd.dk/media/566...
https://www.ddd.dk/media/5662/dvk_epidemiologisk-udredning_ny.pdf
22/21">https://www.ddd.dk/media/566...
Addition: @jbloom_lab& #39;s excellent work shows that Y453F increases affinity of the RBD for human ACE2. Worth noting again, though, that this mutation seems to have arisen multiple times in mink in NL, since April, & is not commonly found in humans.
https://twitter.com/jbloom_lab/status/1324139464818479105
23/21">https://twitter.com/jbloom_la...
https://twitter.com/jbloom_lab/status/1324139464818479105
23/21">https://twitter.com/jbloom_la...
From @ProfJLundgren at University Copenhagen, a list of the 4 mutations found in the Danish sequences.
H69del/V70del & Y453F correspond to the deletion (notated in a different fashion) & mutation covered above & given in the previous reports.
https://twitter.com/ProfJLundgren/status/1324361001869971457
24/21">https://twitter.com/ProfJLund...
H69del/V70del & Y453F correspond to the deletion (notated in a different fashion) & mutation covered above & given in the previous reports.
https://twitter.com/ProfJLundgren/status/1324361001869971457
24/21">https://twitter.com/ProfJLund...
I692V, M1229I were not in the previous reports. Also worth noting that N439K (which I cover above) is not mentioned.
I692V is not present in the sequences we have.
M1229I is found in mostly in Europe - mainly Czech Republic.
https://nextstrain.org/groups/neherlab/ncov/europe?c=gt-S_1229
25/21">https://nextstrain.org/groups/ne...
I692V is not present in the sequences we have.
M1229I is found in mostly in Europe - mainly Czech Republic.
https://nextstrain.org/groups/neherlab/ncov/europe?c=gt-S_1229
25/21">https://nextstrain.org/groups/ne...
Though we see 3 of these mutations (H69del/V70del, Y453F, M1229I) in public data, it seems they usually fall into separate clusters - we haven& #39;t seen them together on the same sequence. Note this isn& #39;t automatically interesting or meaningful.
26/21
26/21
I& #39;m not a structural biologist or virologist. I don& #39;t know of any work on I692V or M1229I, but they are outside of the receptor binding domain (RBD). Here& #39;s a very basic view of where 692 lies (RBD is at the top). 1229 seems like it& #39;s not part of this structure.
27/21
27/21
Hopefully, other scientists who are more familiar with work on these two other mutations and/or any knowledge of what could be gleaned from their positions can weigh in a bit more on what these mutations may/may not mean.
28/21
28/21
@jbloom_lab again provides some great information: Y453F (the mutation seen in other mink in NL, which seems to increase ACE2 binding affinity) reportedly is an escape mutation for an antibody used in the Regeneron cocktail.
https://twitter.com/jbloom_lab/status/1324409358944407554
29/21">https://twitter.com/jbloom_la...
https://twitter.com/jbloom_lab/status/1324409358944407554
29/21">https://twitter.com/jbloom_la...
To boost: @jbloom_lab gives some excellent clarifying answers/information about one of the mutations - read on here:
https://twitter.com/jbloom_lab/status/1324453792432050176
30/21">https://twitter.com/jbloom_la...
https://twitter.com/jbloom_lab/status/1324453792432050176
30/21">https://twitter.com/jbloom_la...
Boosting: @jpglmrodrigues& #39;s great structural work suggests that indeed, the Y453F mutation may help the virus infect mink. Mink have a slightly different ACE2 than humans; this mutation may help binding easier. It may not impact infection in humans
https://twitter.com/jpglmrodrigues/status/1324514230389805057
31/21">https://twitter.com/jpglmrodr...
https://twitter.com/jpglmrodrigues/status/1324514230389805057
31/21">https://twitter.com/jpglmrodr...
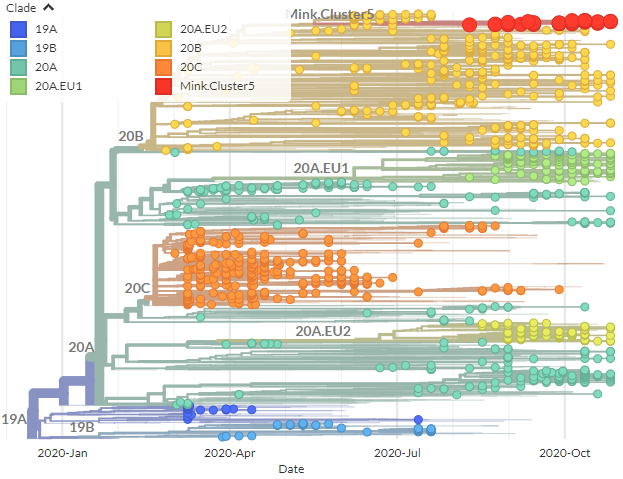
>6000 sequences from Denmark were submitted to @GISAID last night, & we& #39;ve rapidly included them in @richardneher Lab @nextstrain Europe + country builds.
The new Danish seqs (red+orange) cover most of 2020, including very recently. All are human.
https://nextstrain.org/groups/neherlab/ncov/denmark?c=recency&f_country=Denmark
32/21">https://nextstrain.org/groups/ne...
The new Danish seqs (red+orange) cover most of 2020, including very recently. All are human.
https://nextstrain.org/groups/neherlab/ncov/denmark?c=recency&f_country=Denmark
32/21">https://nextstrain.org/groups/ne...
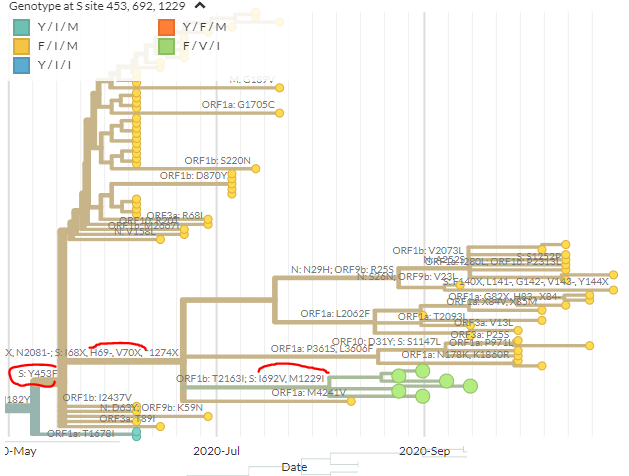
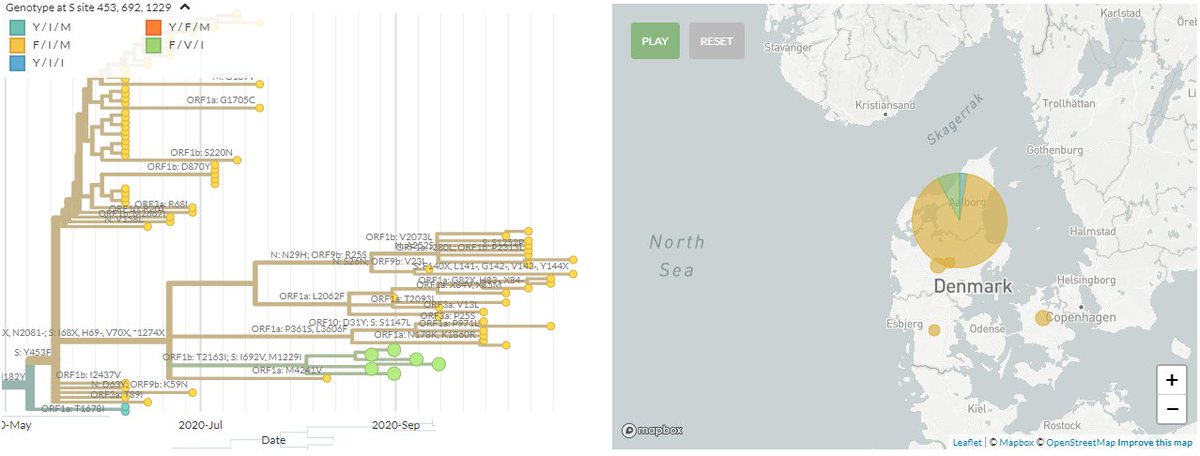
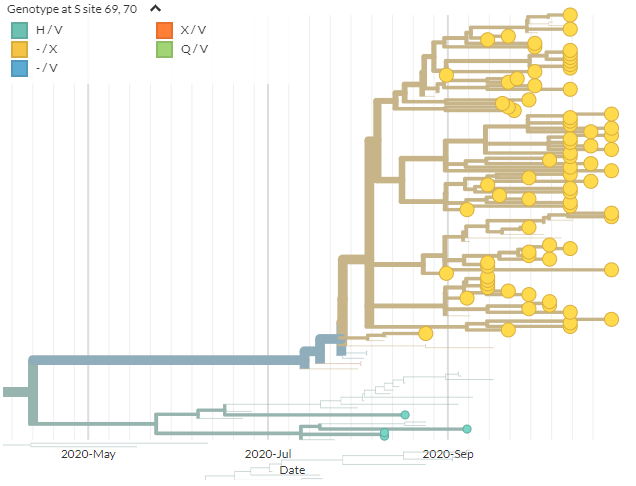
From the mutations reported by SSI, we can zoom in to the & #39;cluster 5& #39; samples that are causing concern (green). In spike, these have the 69/70 deletion, 453F, & 692V, 1229I (marked in red). Divergence view shown on right. As expected, seqs cluster.
https://nextstrain.org/groups/neherlab/ncov/denmark?branchLabel=aa&c=gt-S_453,692,1229&f_country=Denmark&label=mlabel:20B/C15656T&p=grid&r=division
33/21">https://nextstrain.org/groups/ne...
https://nextstrain.org/groups/neherlab/ncov/denmark?branchLabel=aa&c=gt-S_453,692,1229&f_country=Denmark&label=mlabel:20B/C15656T&p=grid&r=division
33/21">https://nextstrain.org/groups/ne...
Also as expected, these sequences come from North Jutland (Nordjylland). There are also closely related sequences from the surrounding area, which have 453F (yellow) - some have the deletion (those clustering nearer to the green), some do not.
34/21
34/21
We& #39;ve added a new clade label to the Denmark build to help highlight this cluster (referred to as & #39;cluster 5& #39; by the SSI) and make it easier to find, given the general attention on it right now. Reminder: all samples right now are rom humans.
35/21
https://nextstrain.org/groups/neherlab/ncov/denmark?f_country=Denmark&p=grid&r=division">https://nextstrain.org/groups/ne...
35/21
https://nextstrain.org/groups/neherlab/ncov/denmark?f_country=Denmark&p=grid&r=division">https://nextstrain.org/groups/ne...
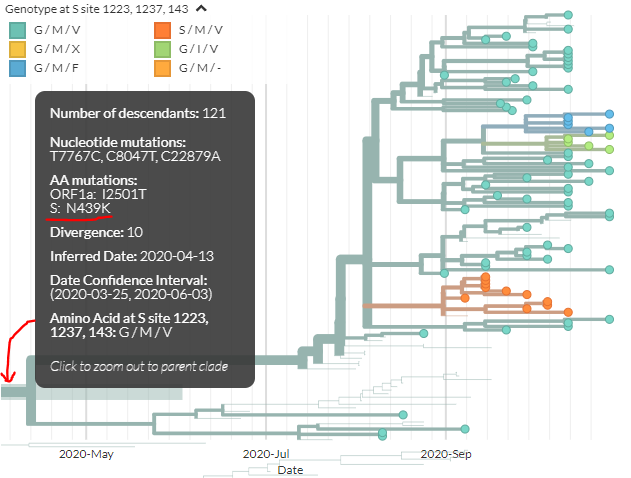
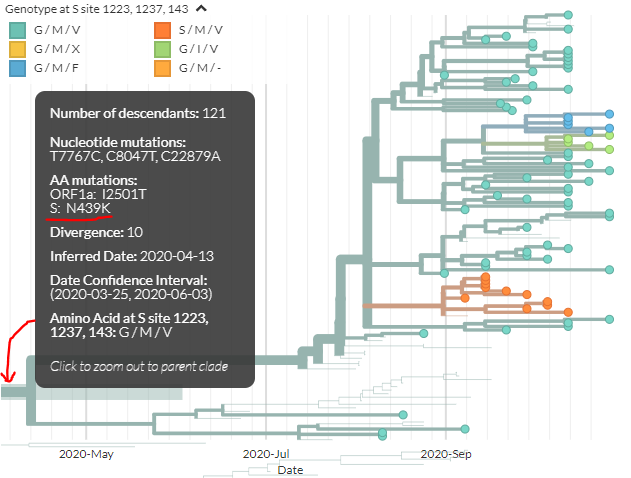
Looking at another, separate cluster that& #39;s gotten attention recently, we can zoom to Danish sequences with N439K (see @robertson_lab& #39;s preprint https://twitter.com/robertson_lab/status/1324470959093460992)
We">https://twitter.com/robertson... can see sequences with multiple mutations in spike: at 143, 1223, & 1237
36/21
https://nextstrain.org/groups/neherlab/ncov/denmark?c=gt-S_1223,1237,143&f_country=Denmark&label=mlabel:20A/G12988T&p=grid&r=division">https://nextstrain.org/groups/ne...
We">https://twitter.com/robertson... can see sequences with multiple mutations in spike: at 143, 1223, & 1237
36/21
https://nextstrain.org/groups/neherlab/ncov/denmark?c=gt-S_1223,1237,143&f_country=Denmark&label=mlabel:20A/G12988T&p=grid&r=division">https://nextstrain.org/groups/ne...
As with 4 out of 5 mutations described in the & #39;cluster 5& #39; seqs, these mutations (apart from 439) aren& #39;t in the receptor binding domain & it& #39;s not clear they would impact antibody binding or a vaccine. This just illustrates that multiple spike mutations may not be unusual.
37/21
37/21
It is also worth mentioning that, as we noted earlier, the 69/70 deletion shows up in this cluster, too (yellow). (70- showing up as X due to an alignment issue). This means this deletion appears to have arisen independently twice.
38/21
https://nextstrain.org/groups/neherlab/ncov/denmark?c=gt-S_69,70&f_country=Denmark&label=mlabel:20A/G12988T&p=grid&r=division">https://nextstrain.org/groups/ne...
38/21
https://nextstrain.org/groups/neherlab/ncov/denmark?c=gt-S_69,70&f_country=Denmark&label=mlabel:20A/G12988T&p=grid&r=division">https://nextstrain.org/groups/ne...
I& #39;ll be running more mutation-focused builds & will update here, as always, when they are up.
39/21
39/21
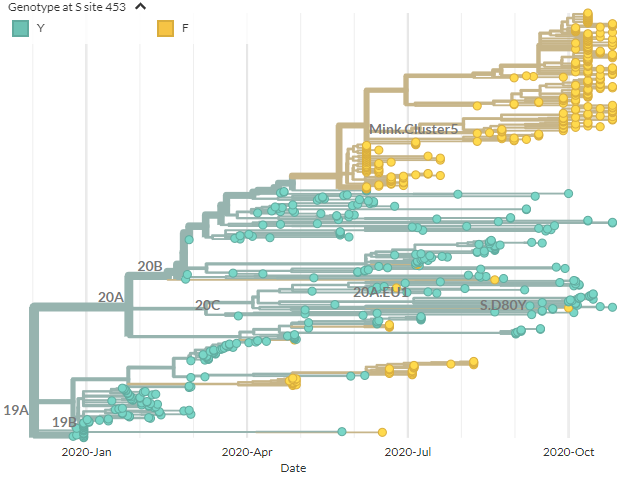
A Y453F-focused build is up. To create this, all sequences with 453F were included, plus closely related sequences & some background.
As we saw yesterday, 453F (yellow) has appeared multiple times, but this now includes Danish seqs (top yellow).
40/21
https://nextstrain.org/groups/neherlab/ncov/S.Y453F?c=gt-S_453">https://nextstrain.org/groups/ne...
As we saw yesterday, 453F (yellow) has appeared multiple times, but this now includes Danish seqs (top yellow).
40/21
https://nextstrain.org/groups/neherlab/ncov/S.Y453F?c=gt-S_453">https://nextstrain.org/groups/ne...
Colouring by host (Neovision & Mustela are mink), we see mink samples from the Netherlands, but are reminded that all Danish samples so far are from humans, though reportedly many came from variants associated with mink farms.
41/21
https://nextstrain.org/groups/neherlab/ncov/S.Y453F?c=host&r=country">https://nextstrain.org/groups/ne...
41/21
https://nextstrain.org/groups/neherlab/ncov/S.Y453F?c=host&r=country">https://nextstrain.org/groups/ne...
The doubling time for this thread is apparently around 50 hours.
(This is a joke https://abs.twimg.com/emoji/v2/... draggable="false" alt="🙃" title="Auf den Kopf gestelltes Gesicht" aria-label="Emoji: Auf den Kopf gestelltes Gesicht">)
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🙃" title="Auf den Kopf gestelltes Gesicht" aria-label="Emoji: Auf den Kopf gestelltes Gesicht">)
42/21
(This is a joke
42/21
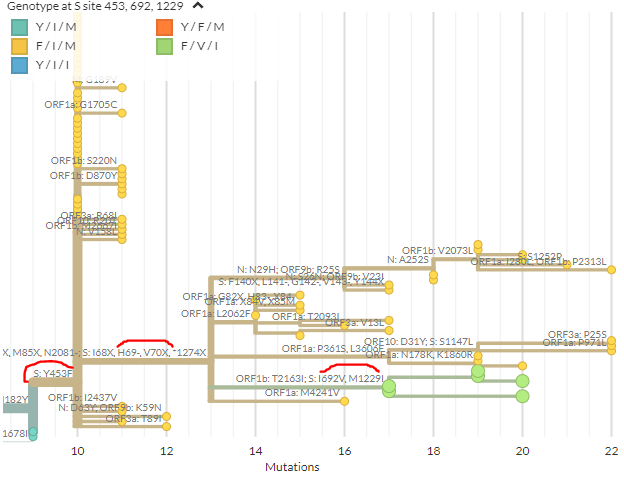
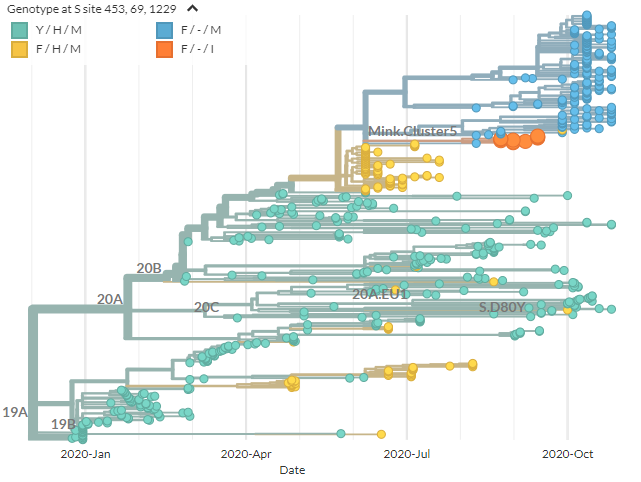
We can add coloring for sites 69 deletion & 1229I (found in the samples of concern). 453F is still yellow, but 453F + deletion is blue, and 453F + deletion & 1229I is orange: this is & #39;cluster 5& #39;.
43/21
https://nextstrain.org/groups/neherlab/ncov/S.Y453F?c=gt-S_453,69,1229">https://nextstrain.org/groups/ne...
43/21
https://nextstrain.org/groups/neherlab/ncov/S.Y453F?c=gt-S_453,69,1229">https://nextstrain.org/groups/ne...
So, we don& #39;t see other samples with 453F & this exact combination of mutations elsewhere - but as I showed earlier, other combinations of spike mutations (without 453F) don& #39;t necessarily seem unusual. It& #39;s unclear if this particular combination is particularly worrisome.
44/21
44/21
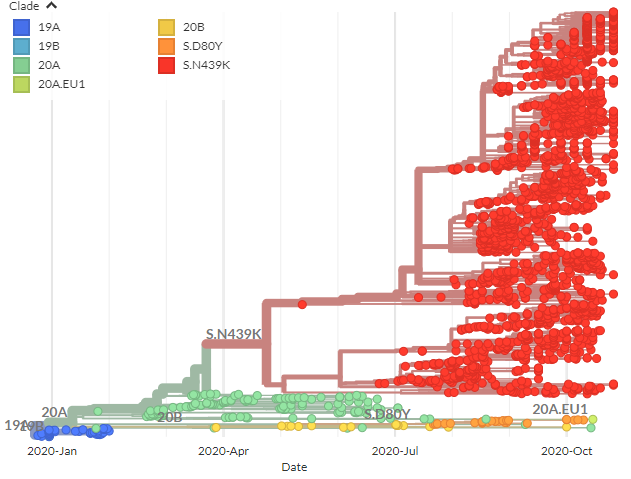
We also have a N439K-focused build. As a reminder, this isn& #39;t a mutation in the & #39;cluster 5& #39; sequences, but is also associated with the double-deletion, is also found in the Danish sequences, & was featured in @robertson_lab& #39;s preprint yesterday.
45/21
https://nextstrain.org/groups/neherlab/ncov/S.N439K">https://nextstrain.org/groups/ne...
45/21
https://nextstrain.org/groups/neherlab/ncov/S.N439K">https://nextstrain.org/groups/ne...
Within the 439K cluster, we can color by the 69 deletion also seen in & #39;cluster 5& #39; sequences (& others with 453F) - here in light grey.
We can see the deletion is ~1/2 of 439K cluster, & found prominently in UK, Denmark, & Switzerland.
46/21
https://nextstrain.org/groups/neherlab/ncov/S.N439K?c=gt-S_69,70&f_region=Europe&label=clade:S.N439K&r=country">https://nextstrain.org/groups/ne...
We can see the deletion is ~1/2 of 439K cluster, & found prominently in UK, Denmark, & Switzerland.
46/21
https://nextstrain.org/groups/neherlab/ncov/S.N439K?c=gt-S_69,70&f_region=Europe&label=clade:S.N439K&r=country">https://nextstrain.org/groups/ne...
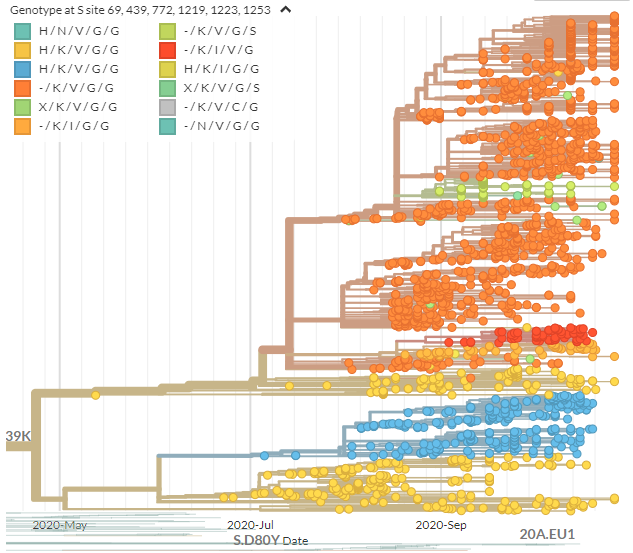
If we color by some of the spike mutations in 439K cluster - there is *a lot* of diversity. This underscores that mutations in spike aren& #39;t uncommon & may not always be interesting.
Scientists mostly focus on those in the receptor binding domain.
47/21
https://nextstrain.org/groups/neherlab/ncov/S.N439K?c=gt-S_69,439,772,1219,1223,1253&f_region=Europe&label=clade:S.N439K&r=country">https://nextstrain.org/groups/ne...
Scientists mostly focus on those in the receptor binding domain.
47/21
https://nextstrain.org/groups/neherlab/ncov/S.N439K?c=gt-S_69,439,772,1219,1223,1253&f_region=Europe&label=clade:S.N439K&r=country">https://nextstrain.org/groups/ne...
Why the focus on 453F & 439K? A quick recap (because I know his is getting long!):
- 453F is one of the mutations in & #39;cluster 5& #39;, linked to mink & of concern to Danish govt. It is in the binding domain
- 439K is not in & #39;cluster 5& #39; but is in Danish seqs & also has attention
48/21
- 453F is one of the mutations in & #39;cluster 5& #39;, linked to mink & of concern to Danish govt. It is in the binding domain
- 439K is not in & #39;cluster 5& #39; but is in Danish seqs & also has attention
48/21
453F (in cluster 5) is also seen in other mink in NL.
It seems to increase human ACE2 binding & is an escape mutation for an antibody used in the Regeneron cocktail ( @jbloom_lab).
It may be an adaptation to mink ACE2.
49/21
https://twitter.com/firefoxx66/status/1324412434203987970">https://twitter.com/firefoxx6... https://twitter.com/firefoxx66/status/1324630245039869953">https://twitter.com/firefoxx6...
It seems to increase human ACE2 binding & is an escape mutation for an antibody used in the Regeneron cocktail ( @jbloom_lab).
It may be an adaptation to mink ACE2.
49/21
https://twitter.com/firefoxx66/status/1324412434203987970">https://twitter.com/firefoxx6... https://twitter.com/firefoxx66/status/1324630245039869953">https://twitter.com/firefoxx6...
439K (also found with the 60/70 deletion, & some Danish seqs have both, though found in other countries too) also seems to decrease antibody binding:
50/21
https://twitter.com/firefoxx66/status/1324095187510042627">https://twitter.com/firefoxx6... https://twitter.com/robertson_lab/status/1324470959093460992">https://twitter.com/robertson...
50/21
https://twitter.com/firefoxx66/status/1324095187510042627">https://twitter.com/firefoxx6... https://twitter.com/robertson_lab/status/1324470959093460992">https://twitter.com/robertson...
From watching the Danish Govt/SSI press conference today, it appears the cluster 5 variant, with all 4 mutations was grown in a lab & tested against recovered patient neutralizing antibodies, where it showed weaker sensitivity than other variants
51/21
https://www.youtube.com/watch?v=zpisnBMtBEI&feature=youtu.be&ab_channel=InternationalPressCentreIPC">https://www.youtube.com/watch... https://www.youtube.com/watch...
51/21
https://www.youtube.com/watch?v=zpisnBMtBEI&feature=youtu.be&ab_channel=InternationalPressCentreIPC">https://www.youtube.com/watch... https://www.youtube.com/watch...
This may explain why we do not yet have information about which mutation exactly the SSI/Danish govt find most concerning, or which antibodies specifically aren& #39;t as sensitive.
Alternatively, this information could be known, but not yet released.
52/21
Alternatively, this information could be known, but not yet released.
52/21
If the first, then it& #39;s possible the reduced sensitivity is from the 453F mutation, which seems to be associated with mink, & is known to be an escape for at least one antibody. This mutation is in mink in NL, though not seen in human seqs.
53/21 https://twitter.com/jbloom_lab/status/1324409358944407554">https://twitter.com/jbloom_la...
53/21 https://twitter.com/jbloom_lab/status/1324409358944407554">https://twitter.com/jbloom_la...
From the press conference, it appears initial announcements that the mutation was a threat to vaccines might be too blunt: it seems this is a possibility, and clearly an impoartant one to investigate, but far from a given. More research is needed, SSI scientists say.
54/21
54/21
The importance of open & clear communication in #SARSCoV2 science reporting is seen here. Open sharing with larger community might also mean we had more answers at this point about which mutation(s) are most concerning.
55/21
55/21
The serious decision to cull all mink in Denmark is not only based on the mutations, but also larger concerns around containing transmission, risk of mutation, & risk of a long-term animal reservoir. With or without cluster 5, that may be the best decision for Denmark.
56/21
56/21
(I will not weigh in on this more directly)
57/21
57/21

 Read on Twitter
Read on Twitter



 https://www.nature.com/articles/..." title="N439K has been reported to confer partial resistance to some antibodies.SSI may have found similar outcomes in their research, if the variant they characterise by the deletions carries N439K, as almost all those we see do. https://elifesciences.org/articles/... href=" https://www.nature.com/articles/s41586-020-2852-111/21">https://www.nature.com/articles/..." class="img-responsive" style="max-width:100%;"/>
https://www.nature.com/articles/..." title="N439K has been reported to confer partial resistance to some antibodies.SSI may have found similar outcomes in their research, if the variant they characterise by the deletions carries N439K, as almost all those we see do. https://elifesciences.org/articles/... href=" https://www.nature.com/articles/s41586-020-2852-111/21">https://www.nature.com/articles/..." class="img-responsive" style="max-width:100%;"/>