and Chris Clarkson ( @c_s_clarkson), Eleanor Drury, Daniel Mead, Jim Stalker, Dushyanth Jyothi, Cinzia Malangone, Mara Lawniczak ( @marakat) and Dominic Kwiatkowski from @sangerinstitute.
Anna Jeffreys, Christina Hubbart, Kate Rowlands and Kirk Rockett from @HumanGeneticsOx
Alistair Miles ( @alimanfoo), Nick Harding ( @nick_harding), Vikki Simpson, and Christa Henrichs from @malariagenomics ...
Alistair Miles ( @alimanfoo), Nick Harding ( @nick_harding), Vikki Simpson, and Christa Henrichs from @malariagenomics ...
Maryam Kamali from Tarbiat Modares University
Dan Schrider @SamurSciCop from @UNC.
Andy Kern ( @pastramimachine) and CJ Battey ( @cj_battey) from @uoregon
David Weetman, Eric Lucas, Alison Isaacs and Martin Donnelly ( @MartinDonnelly8) from @LSTMvector ...
Dan Schrider @SamurSciCop from @UNC.
Andy Kern ( @pastramimachine) and CJ Battey ( @cj_battey) from @uoregon
David Weetman, Eric Lucas, Alison Isaacs and Martin Donnelly ( @MartinDonnelly8) from @LSTMvector ...
Michelle Riehle ( @mmreally1) from @MedicalCollege
Ken Vernick ( @KenVernick) from @institutpasteur
Brad White ( @bjourdanwhite) from @UCRentomology
Craig Wilding ( @Craig_Wilding) from @LJMU
Igor Sharakhov ( @igorshar) from @VT_Entomology
...
Ken Vernick ( @KenVernick) from @institutpasteur
Brad White ( @bjourdanwhite) from @UCRentomology
Craig Wilding ( @Craig_Wilding) from @LJMU
Igor Sharakhov ( @igorshar) from @VT_Entomology
...
Kyanne Rohatgi and Nora Besansky from @ndeckinstitute.
Beniamino Caputo ( @BeniaminoCaputo) and Alessandra della Torre from @SapienzaRoma
Charles Godfray from @OxZooDept
Matt Hahn ( @3rdreviewer) from @IUBiology ...
Beniamino Caputo ( @BeniaminoCaputo) and Alessandra della Torre from @SapienzaRoma
Charles Godfray from @OxZooDept
Matt Hahn ( @3rdreviewer) from @IUBiology ...
João Pinto from IHMT
Samantha O& #39;Loughlin and Austin Burt from @imperialcollege
Carlo Costantini ( @costantini_ird), Jorge Amaya-Romero, Michael Fontaine ( @MikaFontaine1) and Diego Ayala ( @d_ayalag) from @MIVEGEC @ird_fr ...
Samantha O& #39;Loughlin and Austin Burt from @imperialcollege
Carlo Costantini ( @costantini_ird), Jorge Amaya-Romero, Michael Fontaine ( @MikaFontaine1) and Diego Ayala ( @d_ayalag) from @MIVEGEC @ird_fr ...
Boubacar Coulibaly from @ComUSTTB
Janet Midega ( @tjmidega), Charles Mbogo ( @cmmbogo) and Philip Bejon ( @PhilipBejon) @KEMRI_Wellcome
Henry Mawejje from IDRC Uganda
João Dinis from INASA ...
Janet Midega ( @tjmidega), Charles Mbogo ( @cmmbogo) and Philip Bejon ( @PhilipBejon) @KEMRI_Wellcome
Henry Mawejje from IDRC Uganda
João Dinis from INASA ...
Jorge Cano ( @jcano111) from @LSHTM
Arlete Troco from Programa Nacional de Controle da Malaria, Angola
Abdoulaye Diabaté ( @Diabate17699096) from @IrssDro
Nohal Elissa from @pasteurMG ...
Arlete Troco from Programa Nacional de Controle da Malaria, Angola
Abdoulaye Diabaté ( @Diabate17699096) from @IrssDro
Nohal Elissa from @pasteurMG ...
Thank you to coauthors:
Davis Nwakanma and Musa Jawara from @mrcunitgambia
John Essandoh ( @JohnBrain9) from @CapeVars
Edi Constant from @CSRS_CIV
Gilbert Le Goff and Vincent Robert from @MIVEGEC @ird_fr ...
Davis Nwakanma and Musa Jawara from @mrcunitgambia
John Essandoh ( @JohnBrain9) from @CapeVars
Edi Constant from @CSRS_CIV
Gilbert Le Goff and Vincent Robert from @MIVEGEC @ird_fr ...
Ag1000G data will hopefully accelerate the development of new vector control tools. As an example, we built a new map of viable gene drive targets in the doublesex gene.
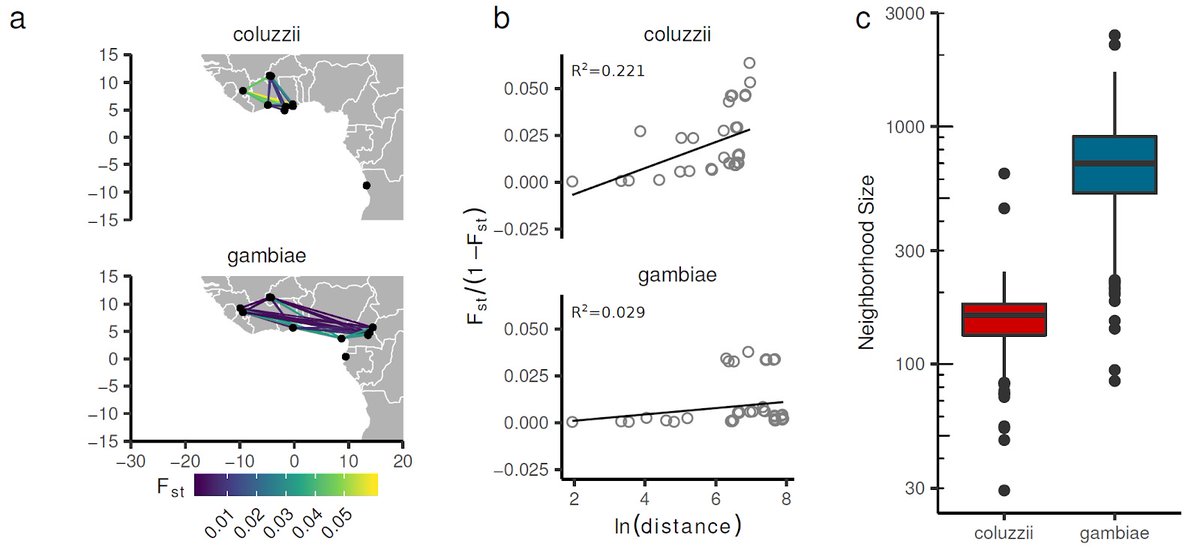
We studied two mosquito species, An. gambiae and An. coluzzii. Still so many unanswered questions about how these malaria vector species are different from each other. But we did find evidence for differences in gene flow and migration behaviour. Much more work needed https://abs.twimg.com/emoji/v2/... draggable="false" alt="🤯" title="Explodierender Kopf" aria-label="Emoji: Explodierender Kopf">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🤯" title="Explodierender Kopf" aria-label="Emoji: Explodierender Kopf"> https://abs.twimg.com/emoji/v2/... draggable="false" alt="💪" title="Angespannter Bizeps" aria-label="Emoji: Angespannter Bizeps">...
https://abs.twimg.com/emoji/v2/... draggable="false" alt="💪" title="Angespannter Bizeps" aria-label="Emoji: Angespannter Bizeps">...
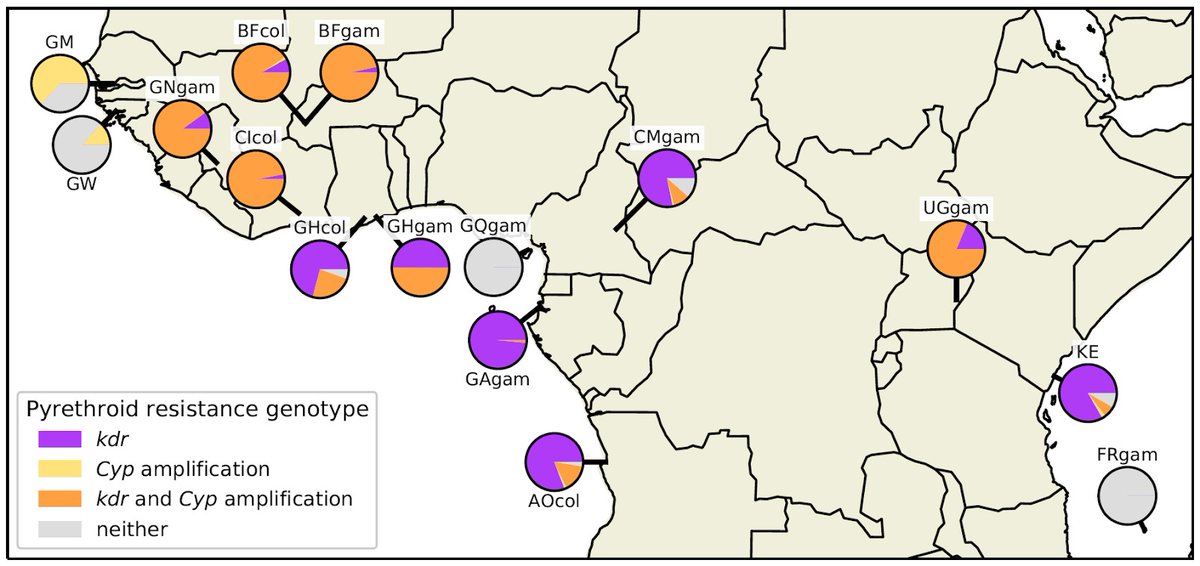
These data contain dozens of new genetic markers for different mechanisms of pyrethroid resistance. This opens up new possibilities for molecular surveillance of insecticide resistance.
We found:
 https://abs.twimg.com/emoji/v2/... draggable="false" alt="☑️" title="Kästchen mit Häkchen" aria-label="Emoji: Kästchen mit Häkchen"> 57 million SNPs,
https://abs.twimg.com/emoji/v2/... draggable="false" alt="☑️" title="Kästchen mit Häkchen" aria-label="Emoji: Kästchen mit Häkchen"> 57 million SNPs,
 https://abs.twimg.com/emoji/v2/... draggable="false" alt="☑️" title="Kästchen mit Häkchen" aria-label="Emoji: Kästchen mit Häkchen"> 31 thousand CNVs,
https://abs.twimg.com/emoji/v2/... draggable="false" alt="☑️" title="Kästchen mit Häkchen" aria-label="Emoji: Kästchen mit Häkchen"> 31 thousand CNVs,
 https://abs.twimg.com/emoji/v2/... draggable="false" alt="☑️" title="Kästchen mit Häkchen" aria-label="Emoji: Kästchen mit Häkchen"> built a new haplotype reference panel.
https://abs.twimg.com/emoji/v2/... draggable="false" alt="☑️" title="Kästchen mit Häkchen" aria-label="Emoji: Kästchen mit Häkchen"> built a new haplotype reference panel.
All data are openly available https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Rückhand Zeigefinger nach unten" aria-label="Emoji: Rückhand Zeigefinger nach unten">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Rückhand Zeigefinger nach unten" aria-label="Emoji: Rückhand Zeigefinger nach unten">
https://www.malariagen.net/resource/27 ">https://www.malariagen.net/resource/...
All data are openly available
https://www.malariagen.net/resource/27 ">https://www.malariagen.net/resource/...
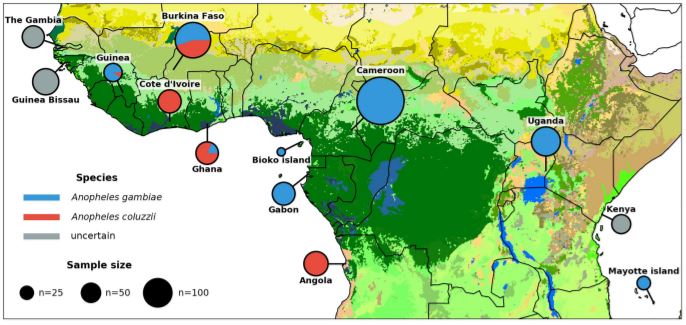
The Ag1000G phase 2 resource has genome variation data from deep sequencing of 1,142 wild-caught mosquitoes from 13 countries...
The Anopheles gambiae 1000 genomes (Ag1000G) project phase 2 paper is out in @genomeresearch  https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Rückhand Zeigefinger nach unten" aria-label="Emoji: Rückhand Zeigefinger nach unten">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Rückhand Zeigefinger nach unten" aria-label="Emoji: Rückhand Zeigefinger nach unten">
https://genome.cshlp.org/content/early/2020/09/25/gr.262790.120.full.pdf+html
Huge">https://genome.cshlp.org/content/e... thank you https://abs.twimg.com/emoji/v2/... draggable="false" alt="🙏" title="Folded hands" aria-label="Emoji: Folded hands">to everyone who helped make this happen. This thread
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🙏" title="Folded hands" aria-label="Emoji: Folded hands">to everyone who helped make this happen. This thread  https://abs.twimg.com/emoji/v2/... draggable="false" alt="🧵" title="Thread" aria-label="Emoji: Thread">gives some paper highlights ...
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🧵" title="Thread" aria-label="Emoji: Thread">gives some paper highlights ...
https://genome.cshlp.org/content/early/2020/09/25/gr.262790.120.full.pdf+html
Huge">https://genome.cshlp.org/content/e... thank you

 Read on Twitter
Read on Twitter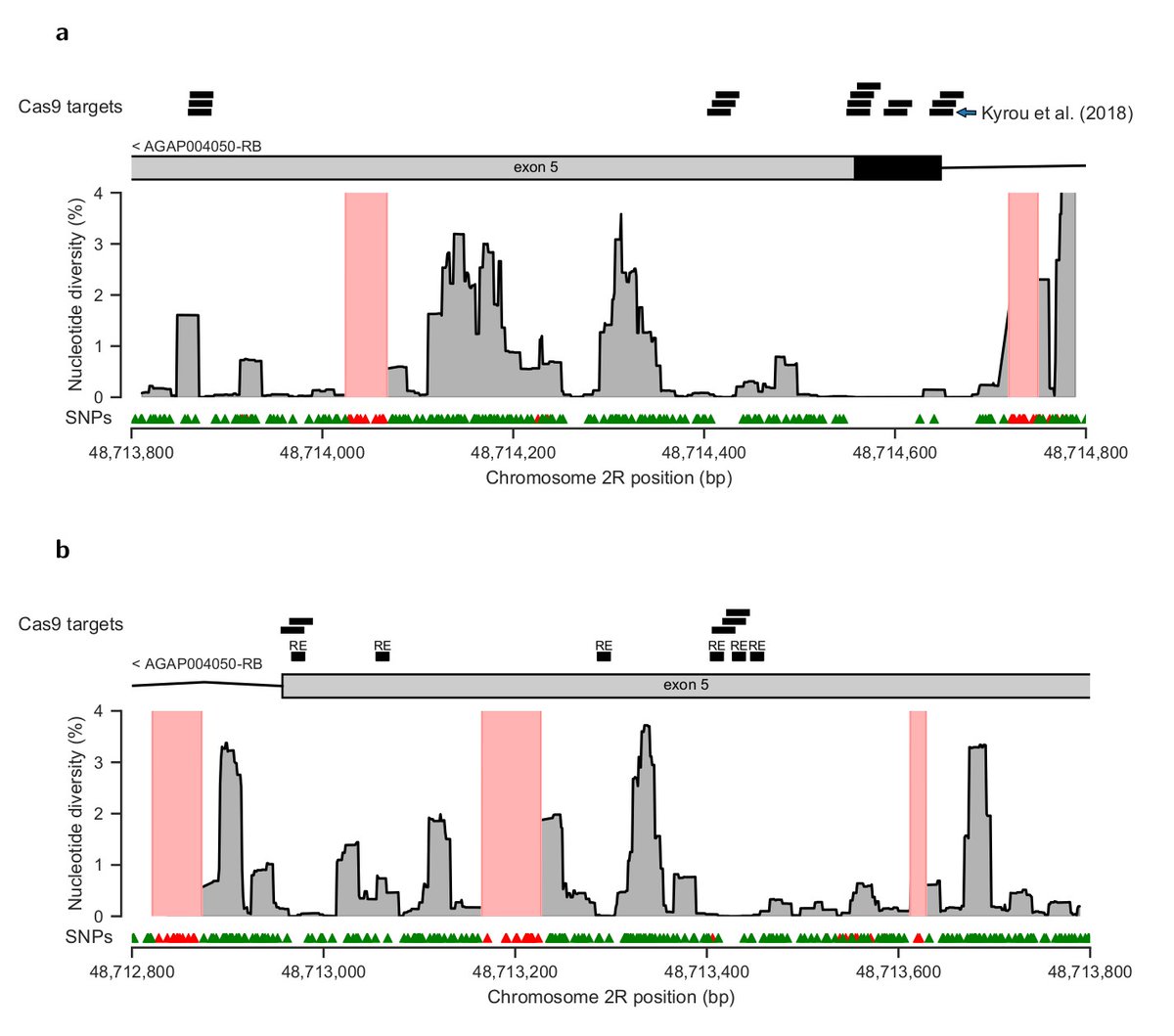
 https://abs.twimg.com/emoji/v2/... draggable="false" alt="💪" title="Angespannter Bizeps" aria-label="Emoji: Angespannter Bizeps">..." title="We studied two mosquito species, An. gambiae and An. coluzzii. Still so many unanswered questions about how these malaria vector species are different from each other. But we did find evidence for differences in gene flow and migration behaviour. Much more work neededhttps://abs.twimg.com/emoji/v2/... draggable="false" alt="🤯" title="Explodierender Kopf" aria-label="Emoji: Explodierender Kopf">https://abs.twimg.com/emoji/v2/... draggable="false" alt="💪" title="Angespannter Bizeps" aria-label="Emoji: Angespannter Bizeps">..." class="img-responsive" style="max-width:100%;"/>
https://abs.twimg.com/emoji/v2/... draggable="false" alt="💪" title="Angespannter Bizeps" aria-label="Emoji: Angespannter Bizeps">..." title="We studied two mosquito species, An. gambiae and An. coluzzii. Still so many unanswered questions about how these malaria vector species are different from each other. But we did find evidence for differences in gene flow and migration behaviour. Much more work neededhttps://abs.twimg.com/emoji/v2/... draggable="false" alt="🤯" title="Explodierender Kopf" aria-label="Emoji: Explodierender Kopf">https://abs.twimg.com/emoji/v2/... draggable="false" alt="💪" title="Angespannter Bizeps" aria-label="Emoji: Angespannter Bizeps">..." class="img-responsive" style="max-width:100%;"/>