I& #39;m very pleased to announce our publication "Whole genome, transcriptome and methylome profiling enhances actionable target discovery in high-risk paediatric cancer" online today in @NatureMedicine https://rdcu.be/b76h9 ">https://rdcu.be/b76h9&quo... Here& #39;s a tweetorial of the main findings 1/22
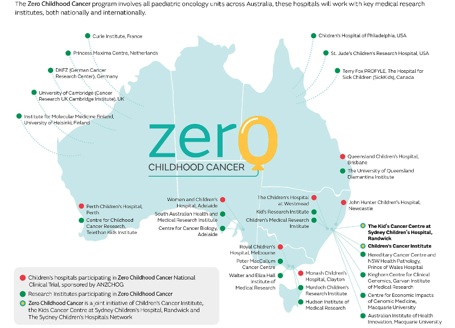
This research was 7 years in the making & is the first major publication from #ZeroChildhoodCancer, a joint initiative of @KidsCancerInst and Kids Cancer Centre @Sydney_Kids. #Zero is now a national precision medicine program, including all 8 hospitals and 23 research partners /2
What value does deciphering the whole genome hold for guiding clinical care of children with high-risk cancer? LOTS. Does it provide more accurate diagnoses, more treatment options, better risk stratification? YES. Can it be done fast enough to have a clinical impact? YES. /3
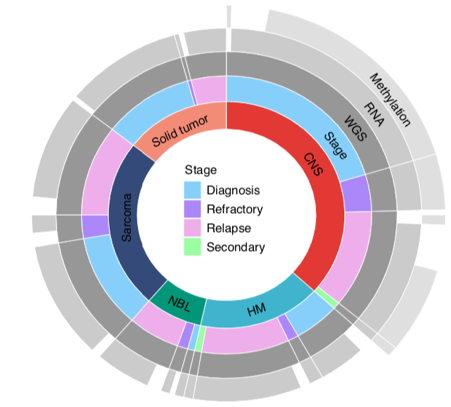
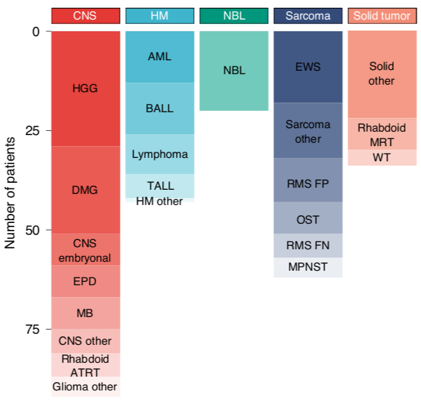
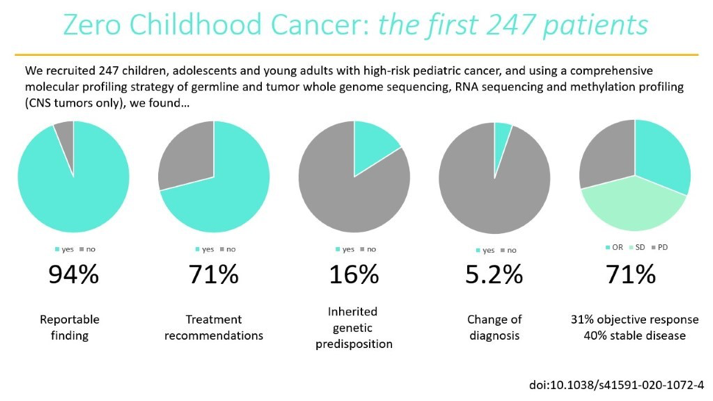
Here, we recruited 247 young people with the most aggressive forms of childhood cancer, with <30% estimated chance of surviving 5 years. The cohort was heterogeneous wrt age, diagnosis & stage. Half were heavily pre-treated. Most had no more treatment options available. /4
We hypothesised that a comprehensive molecular profiling strategy incl. germline (>30x) and tumour (>90x) whole genome sequencing ( #WGS), RNAseq and methylation profiling would resolve the molecular basis of each tumour & have a high clinical impact. /5
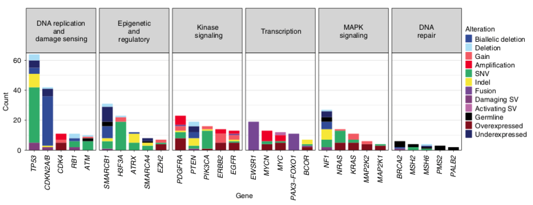
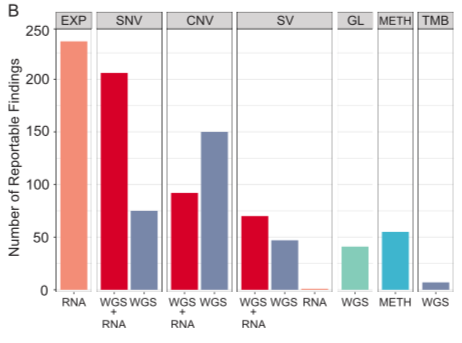
Here we show that #WGS and RNAseq resolve the myriad molecular changes that drive childhood tumour. We found many atypical gene fusions and other driver structural variants that would be missed by standard pathology or more targeted approaches. led by Marie Wong. /6
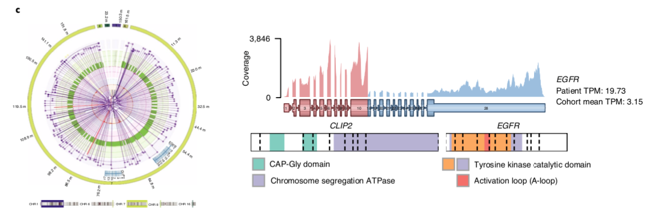
The combination of #WGS and #RNAseq revealed often complex patterns of SV, producing novel forms of activated gene fusions. For example, we identified a novel EGFR activating fusion, recommended 2nd gen EGFRi, having a significant and durable response. /7
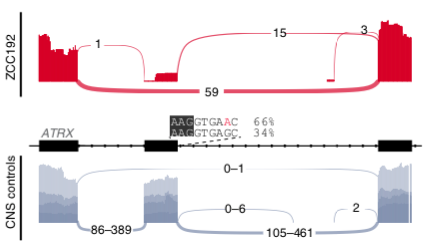
The combo of #WGS+ #RNAseq provided insights into the functional impact of mutations on gene splicing. 9/28 splice-altering variants found by #Introme were at atypical sites & RNAseq revealed exon skipping & whole or partial intron retention. led by @PatSullivann /8
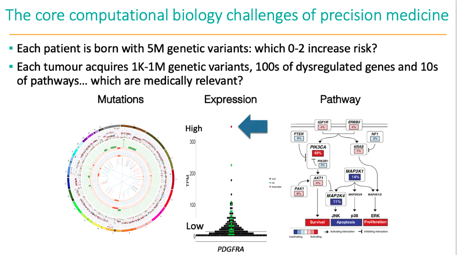
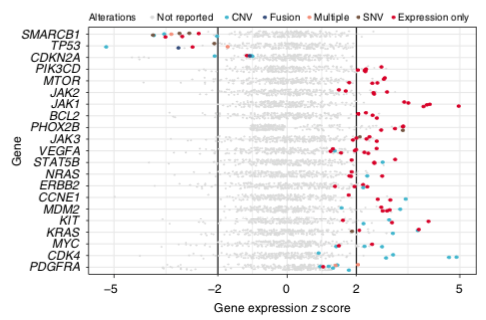
We reported genes with aberrant abundance as potential treatment targets for targeted therapies, even in the absence of explanatory molecular events like copy number amplification. In some cases, this led to significant tumour shrinkage. Led by @cmayoh_bio /9
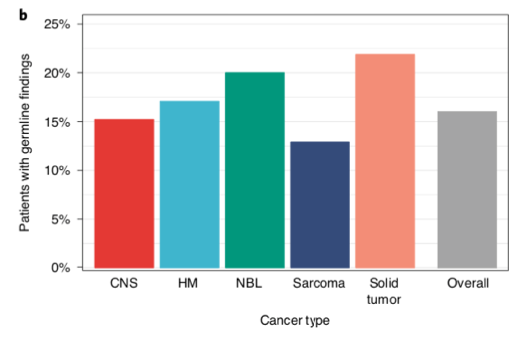
When we focussed on the germline #WGS, we found 16% of patients had a pathogenic cancer predisposition variant; 2/3 of these were not known before #Zero & has big implications for identifying at-risk individuals before they develop cancer. Far higher # than expected. /10
Incorporating somatic features from the tumour, including tumour mutation burden, mutational signatures and somatic second hit mutations informed the pathogenicity assessment of the germline variants. Informatics led by @MarkPinese /11
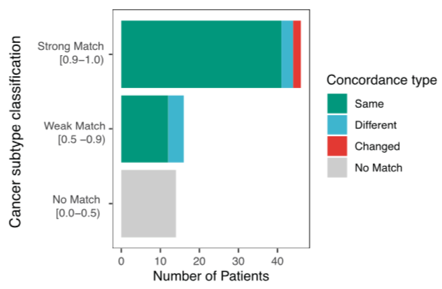
Diagnosing many brain tumours is very challenging. Methylation profiling of tumours and using the MNP classifier ( https://www.molecularneuropathology.org/mnp )">https://www.molecularneuropathology.org/mnp"... helped confirm or change diagnosis in many cases. This is rapidly becoming an essential test. /12
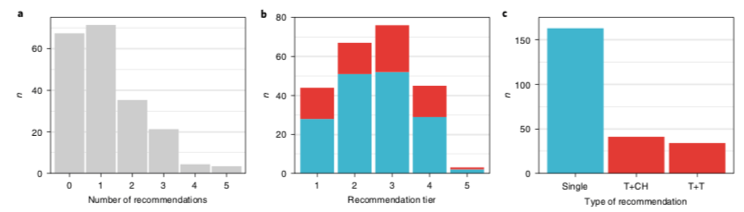
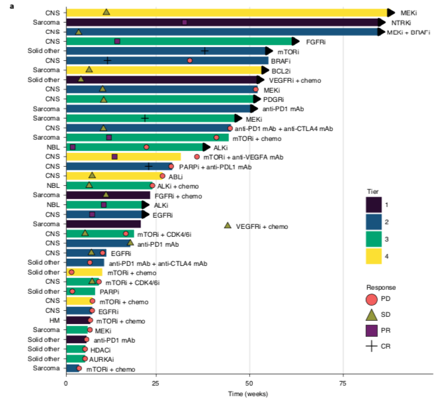
This data is then distilled & presented at a national #MTB. 71% of patients received >=1 treatment recommendation (up to 5), across a range of tiers, 1 highest evidence in the same cancer type, down to a consensus opinion. led by Loretta Lau, Dong Anh Khuong Quang /13
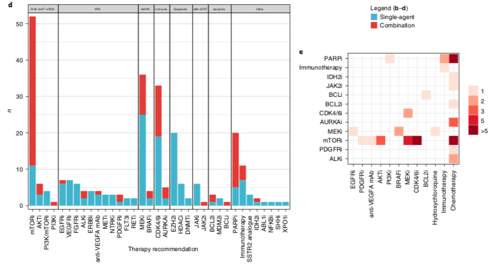
The most common drug targets were in the PI3K/AKT/mTOR pathway, various receptor tyrosine kinases, the MAPK pathway, the cell cycle complex and epigenetic pathways including SWI/SNF/PRC2 chromatin remodelling complex. Drugs were accessed through trials & compassionate access /14
Almost 1/3 of patients received the recommended therapies. 31% had an objective clinical response and 40% had disease stabilisation, many for >24 weeks. The tier/evidence strength of recommendation had little correlation with the clinical response and/or duration of response. /15
The combo of germline & tumour #WGS, #RNAseq and methylation profiling led to far higher than expected numbers of reportable findings & establishing a diagnosis. Only 40% of events were found by both RNA and WGS, and this was far lower earlier in the program - #methodsmatter. /16
The main takehome msg is that #WGS + #RNAseq + methylation profiling is a powerful precision medicine platform for children with high-risk cancer, with substantial clinical impact & can be delivered nationally in a clinical timeframe (avg 7.5 weeks). /17
#ZeroChildhoodCancer would not be possible without the trust of patients and their parents. Enormous thanks to the large and tireless team of scientists, clinicians and leadership team throughout Australia who make this happen. Special thanks to Computational Biology Group! /18
Congratulations to our co-first authors Marie Wong (WGS lead), @cmayoh_bio (RNA lead), Loretta Lau (Precision oncologist lead) for your tireless efforts to develop the analytical tools to deliver #ZeroChildhoodCancer. /19
Thank you to our many industry and pharmaceutical partners for working closely with us to develop the technologies required to deliver this platform. Cutting-edge technologies and drug access are fundamental to the success of this program. /20
Many thanks to @AProfDavidZieg1 (clincial study chair) and Paul Ekert (zero curation team and functional genomics lead) for co-leading this manuscript with me. Thank you to the #Zero leadership team, Michelle Haber, Glenn Marshall, Vanessa Tyrrell. /21
And finally, a sincere thanks to our generous supporters who took a chance, worked tireless to fund-raise, promote awareness & enabled us to develop #ZeroChildhoodCancer. There is much more to do, but today we will celebrate this milestone! /end

 Read on Twitter
Read on Twitter