[Thread] 1/22 Check out our pan-virus analysis! Tremendous teamwork by @bitsoftime @adityarao95 Denis Dermadi, @jiaying_toh, @ladoctorajones @mdonato1138 and @yemlooo with our collaborators @LabHeath @isbsci and @yehudithasin and Yudong He @inflammatix_inc https://medrxiv.org/cgi/content/short/2020.10.02.20205880v1">https://medrxiv.org/cgi/conte...
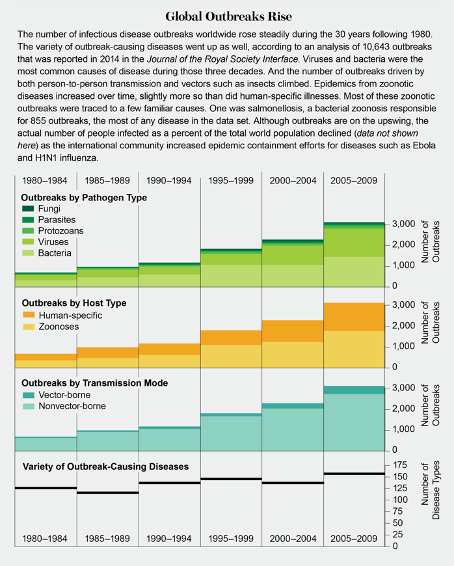
2/ WHY? With the fourth pandemic since 2009, we are in the pandemic era ( https://www.cell.com/cell/fulltext/S0092-8674(20)31012-6),">https://www.cell.com/cell/full... but the number of outbreaks has been on the rise for decades (via Scientific American)
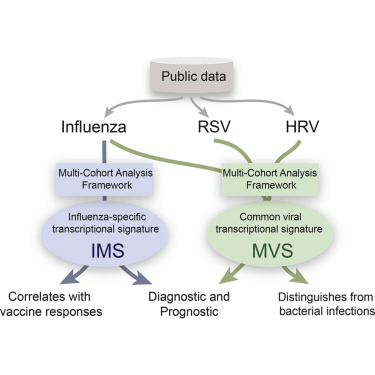
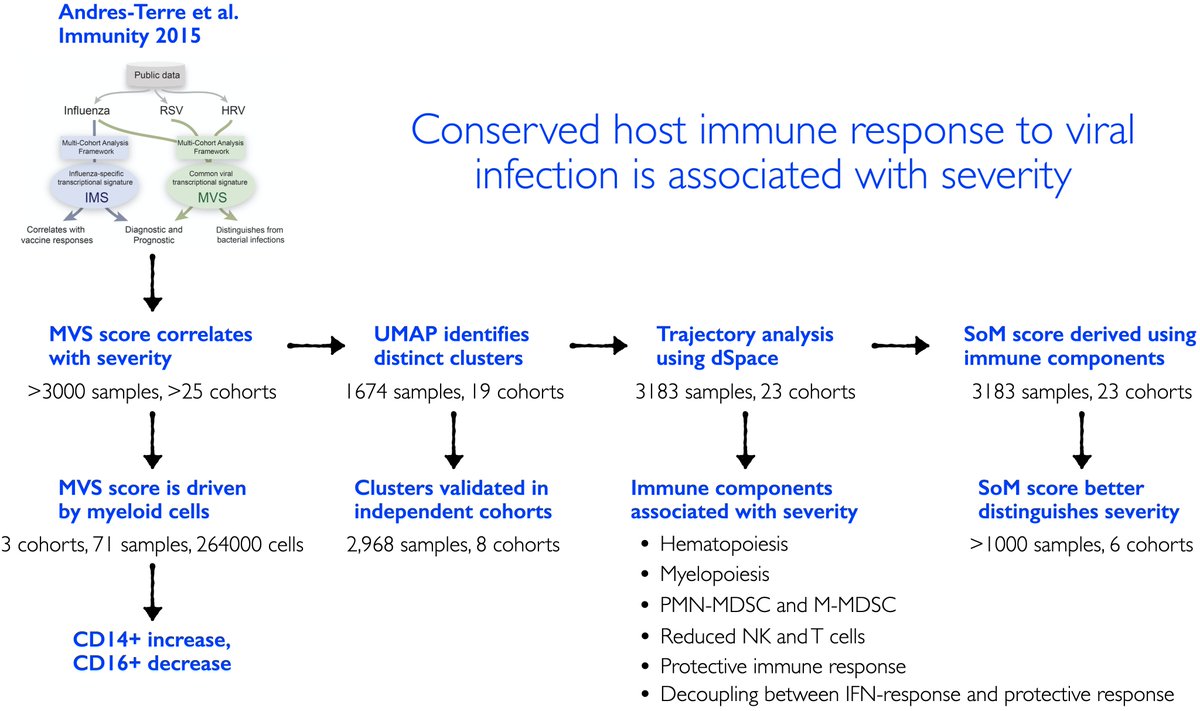
3/ Hypothesis: Our previously described conserved host response to viral infection by @Apuntsdelcami, called meta-virus signature (MVS), could be a generalizable tool in our pandemic preparedness. https://www.cell.com/immunity/fulltext/S1074-7613(15)00455-0">https://www.cell.com/immunity/...
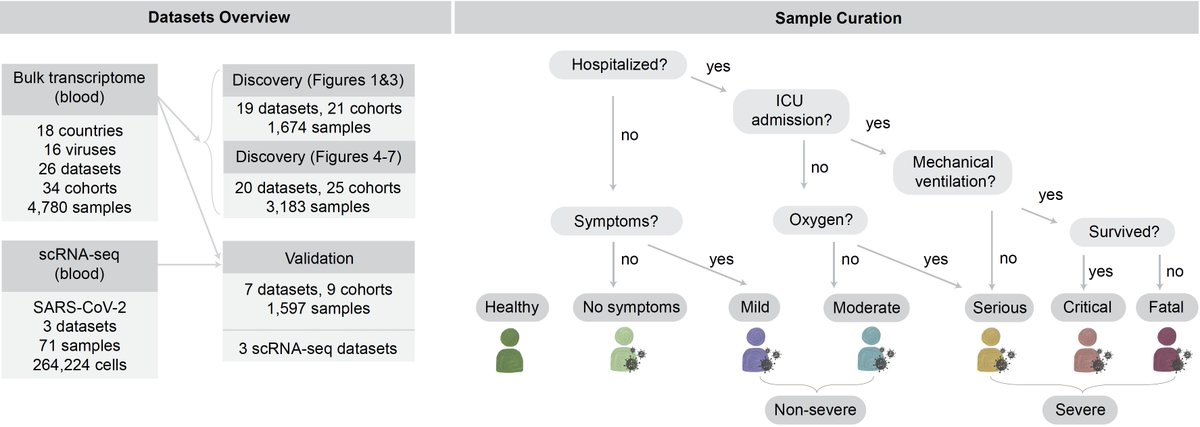
4/ To test this hypothesis, we collected and curated large amount of public data – 4780 bulk transcriptomes in 34 cohorts from 18 countries with one of 16 viruses. We also collected scRNA-seq for 264000 cells from 71 samples across 3 cohorts.
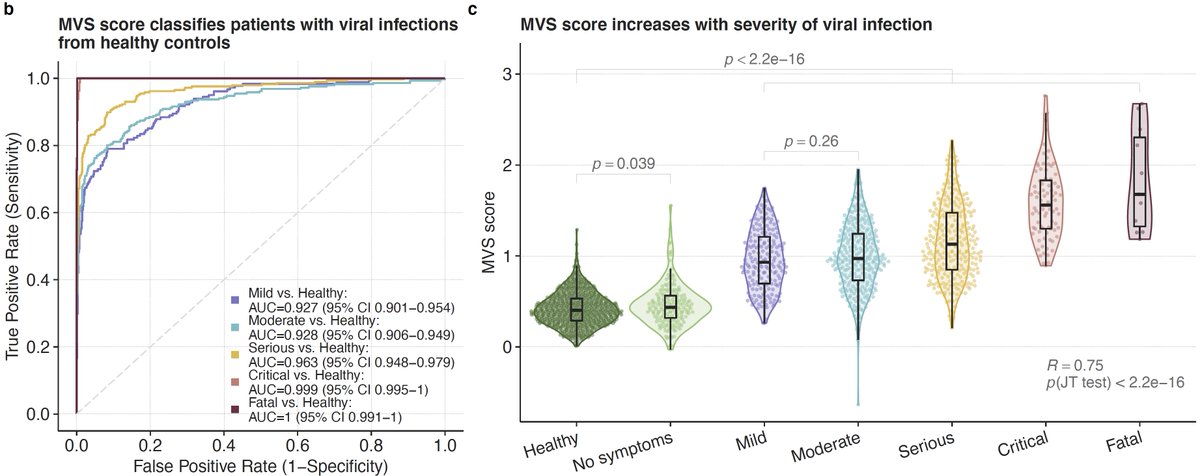
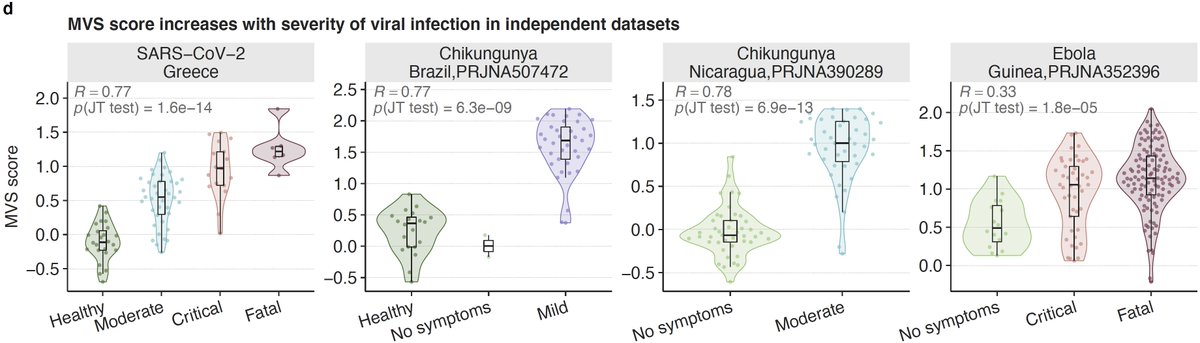
5/ We found that the MVS score, as we described before, is correlated with severity of viral infection irrespective of the type of virus. No difference in those with mild and moderate infection. But will come back that later.
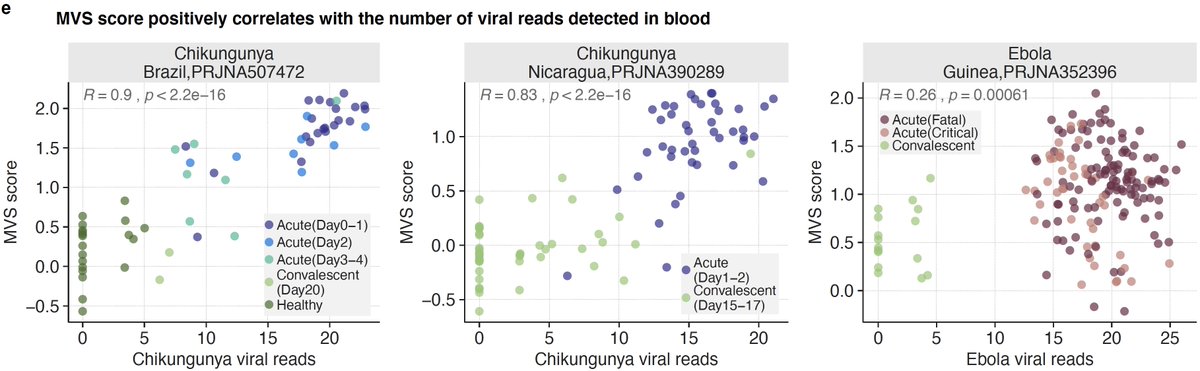
6/ The MVS score is also correlated with severity and the number of viral reads detected in blood in patients with viral infections that were not used in our original paper including SARS-CoV-2, chikungunya, and Ebola (4 countries and 3 viruses).
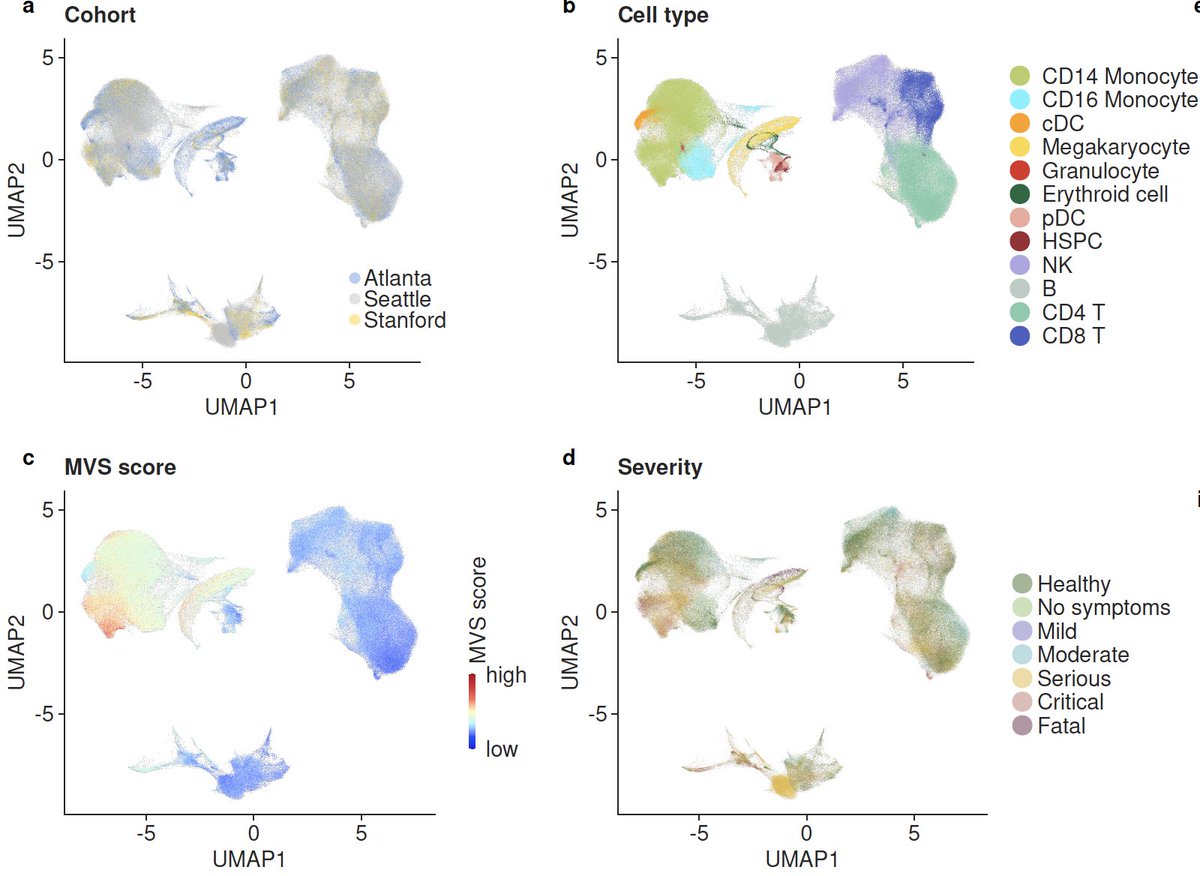
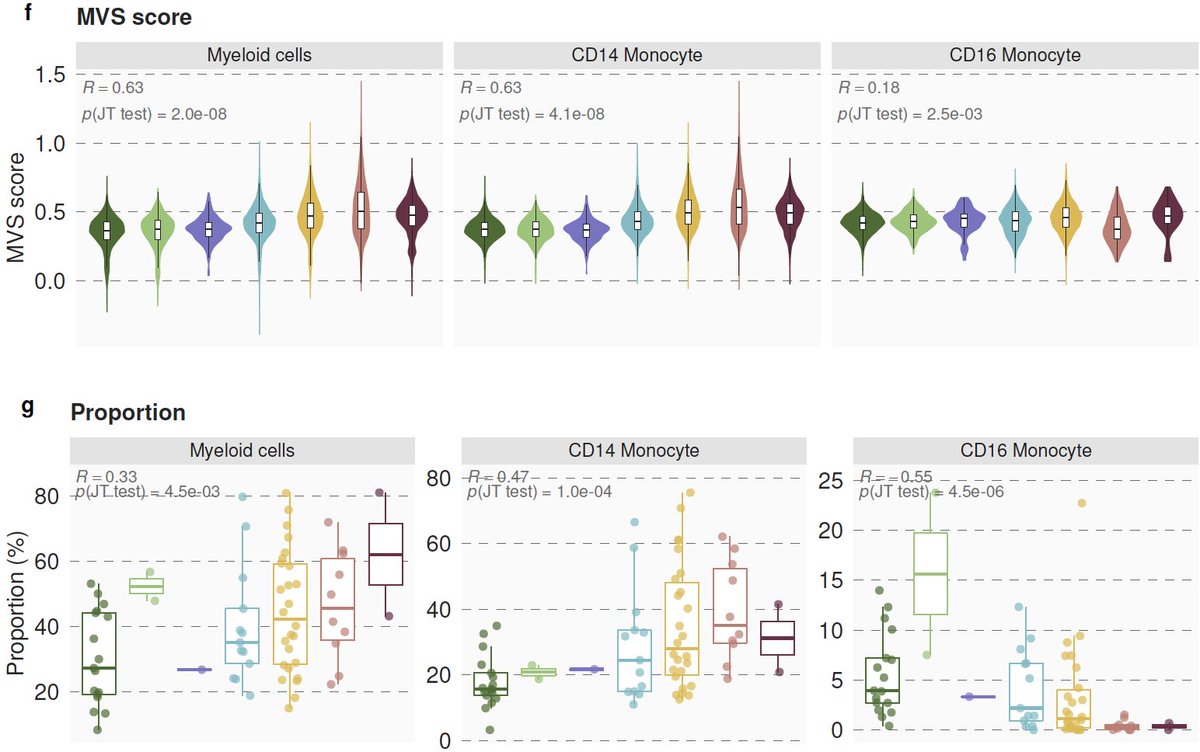
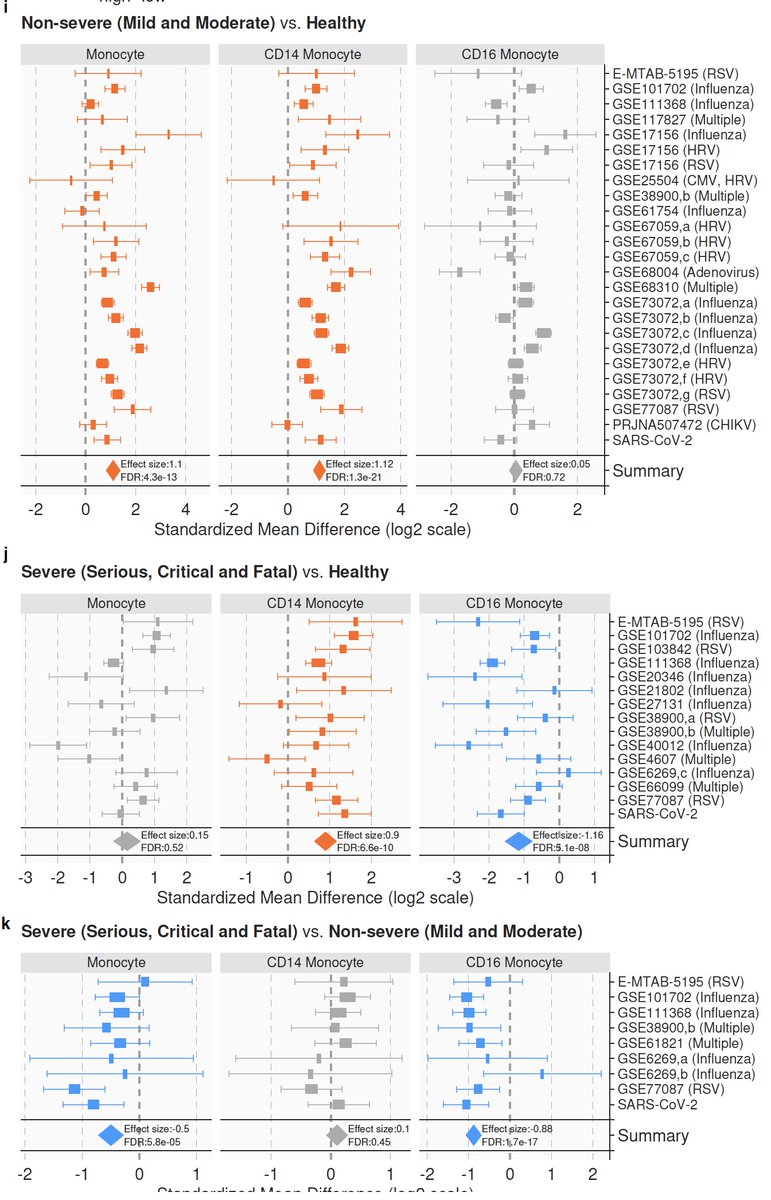
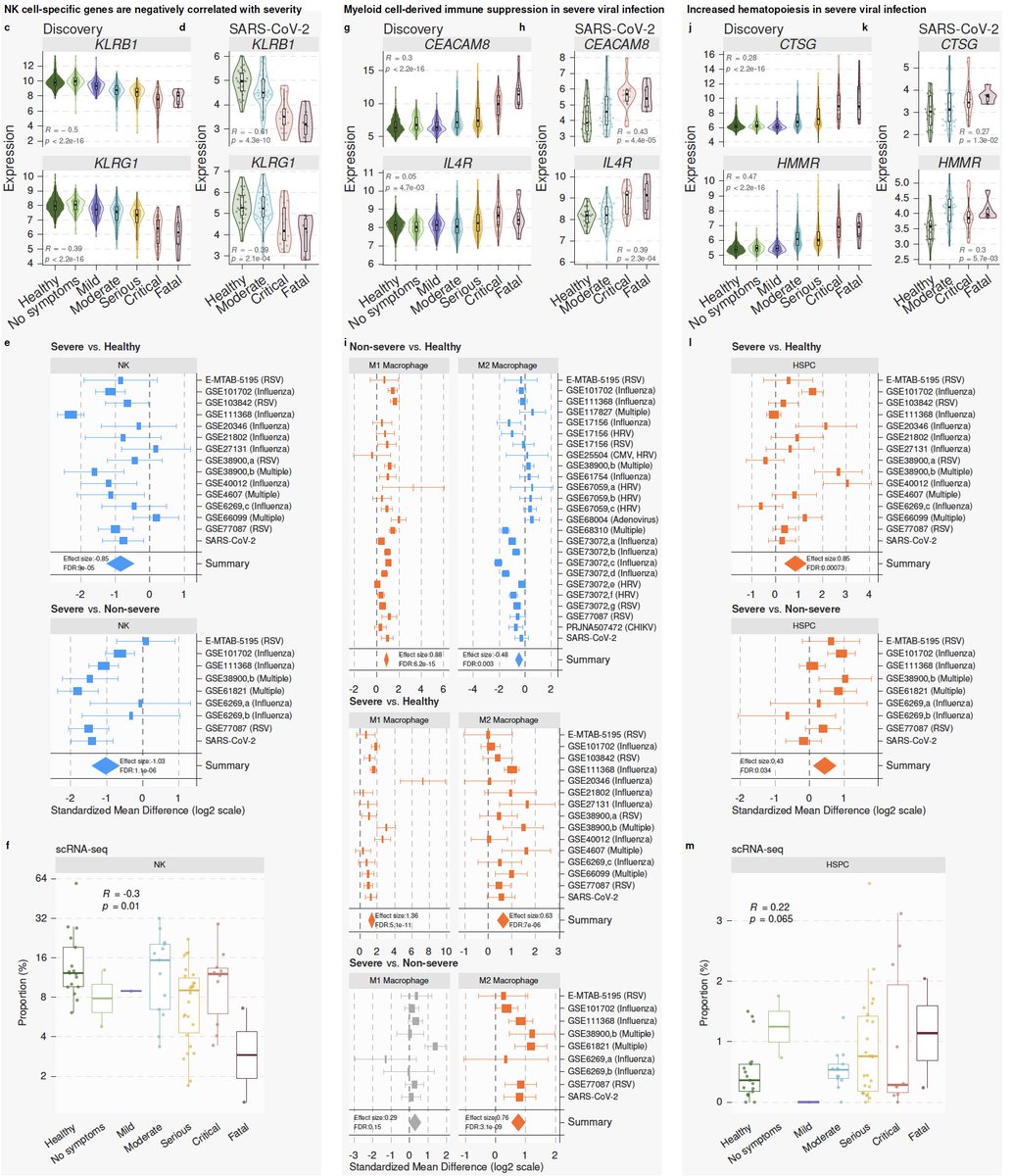
7/ We integrated >264000 scRNA-seq profiles from 3 centers to find the source of the MVS. We found the MVS is dominated by myeloid cells (mostly CD14) and higher in patients with severe outcomes. Proportions of CD14 increase and CD16 decrease with severity.
8/ In silico cellular deconvolution using immunoStates ( https://www.nature.com/articles/s41467-018-07242-6)">https://www.nature.com/articles/... of 4200 samples also finds lower CD16 proportions with increased severity of viral infection. Results are consistent at single-cell and bulk level across all viruses we analyzed.
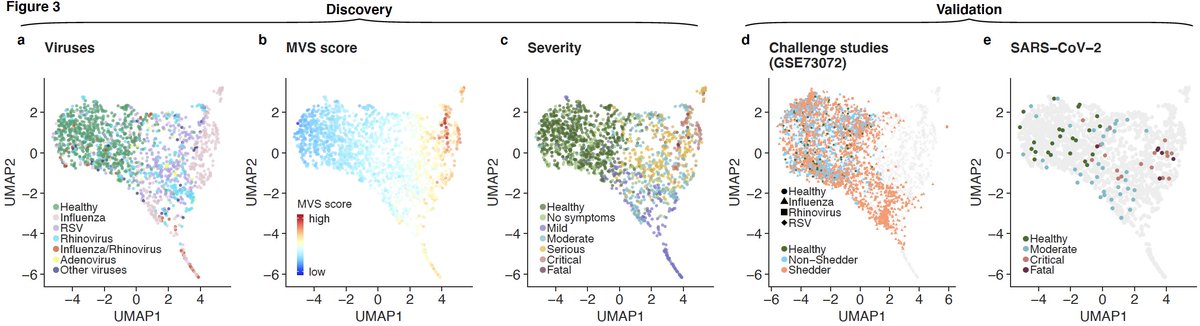
9/ We asked if the MVS distinguishes patients with mild or severe viral infection using bulk transcriptome profiles. UMAP using 1675 samples from 21 cohorts suggests that it does. We validate this observation in 8 independent datasets from 4 viruses including SARS-CoV-2.
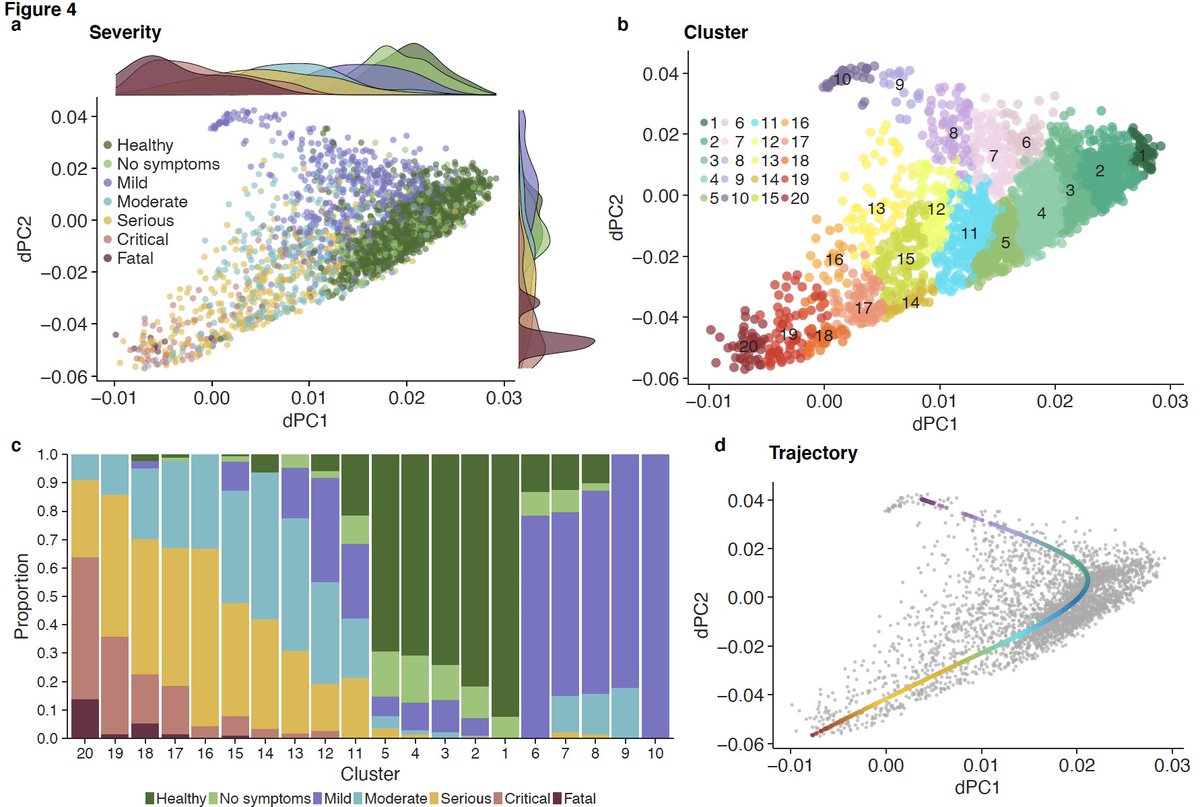
10/ We adapted tSpace, a cellular differentiation trajectory method, and applied to bulk transcriptome profiles to infer trajectory of patients with viral infection. We call it dSpace as in “disease space.”
11/ dSpace shows patients with mild viral infection follow different trajectory than those with severe viral infection irrespective of the infecting virus.
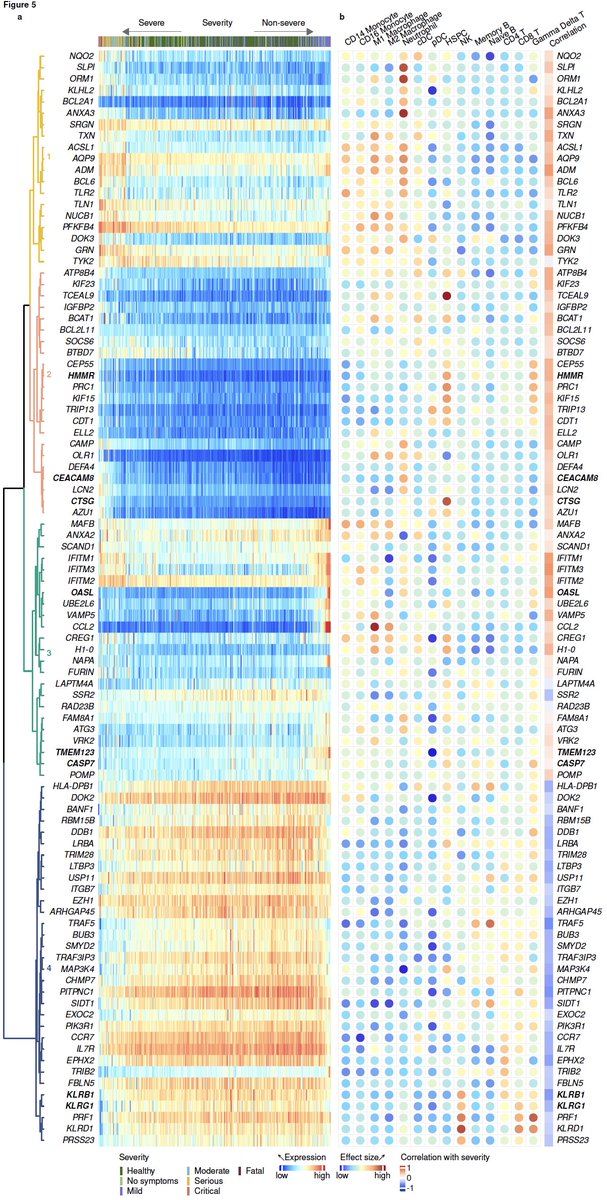
12/ We found 96 genes are significantly associated with the trajectory (left). We also found that genes with positive or negative correlation with severity are biased towards myeloid or lymphoid cells, respectively (right), using MetaSignature database ( https://metasignature.stanford.edu"> https://metasignature.stanford.edu ).
13/ We learned a lot from these 96 genes. They suggest increased hematopoietic progenitors and myeloid-derived suppressor cells, and decreased NK cells with increased severity of viral infection. We validate each of these using bulk and single-cell transcriptome profiles.
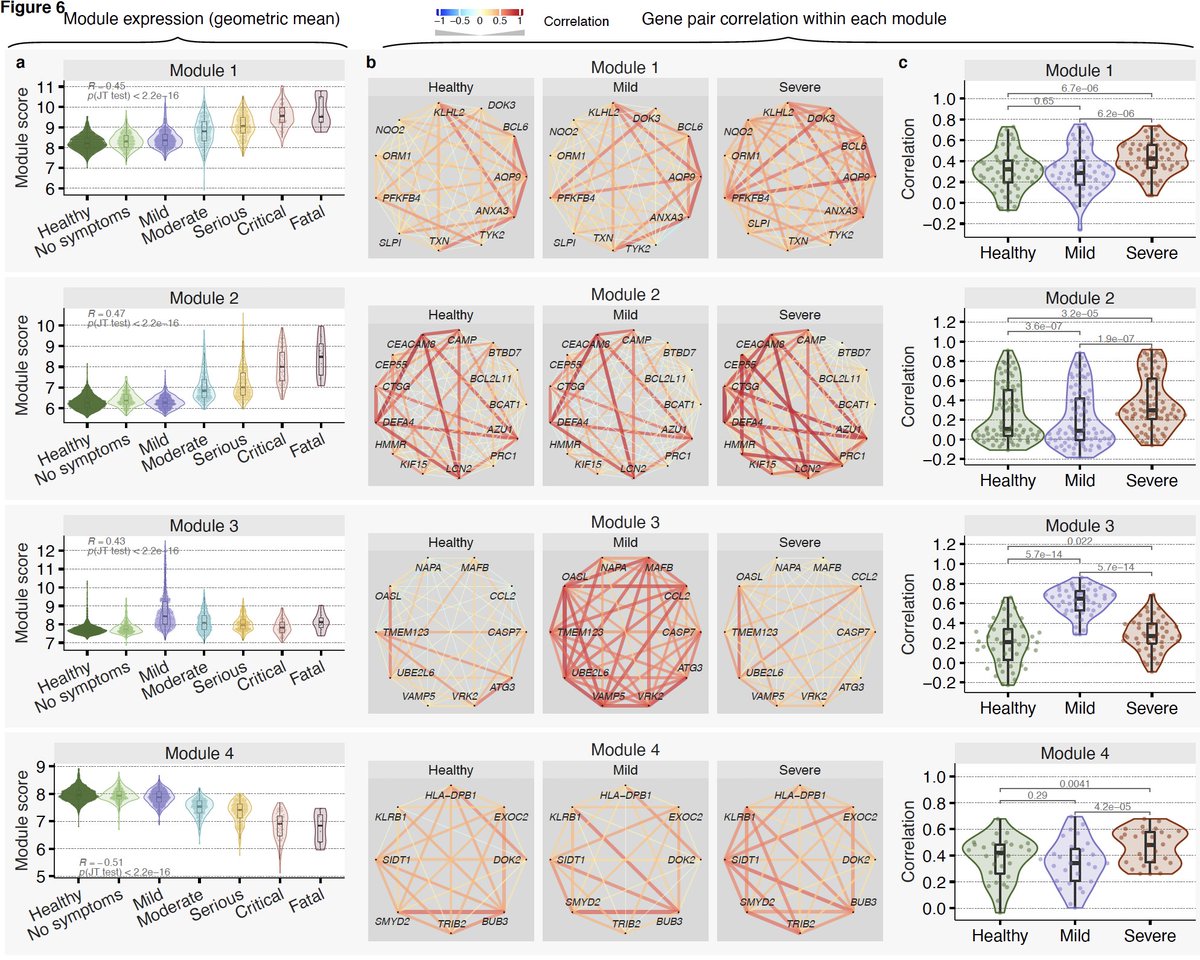
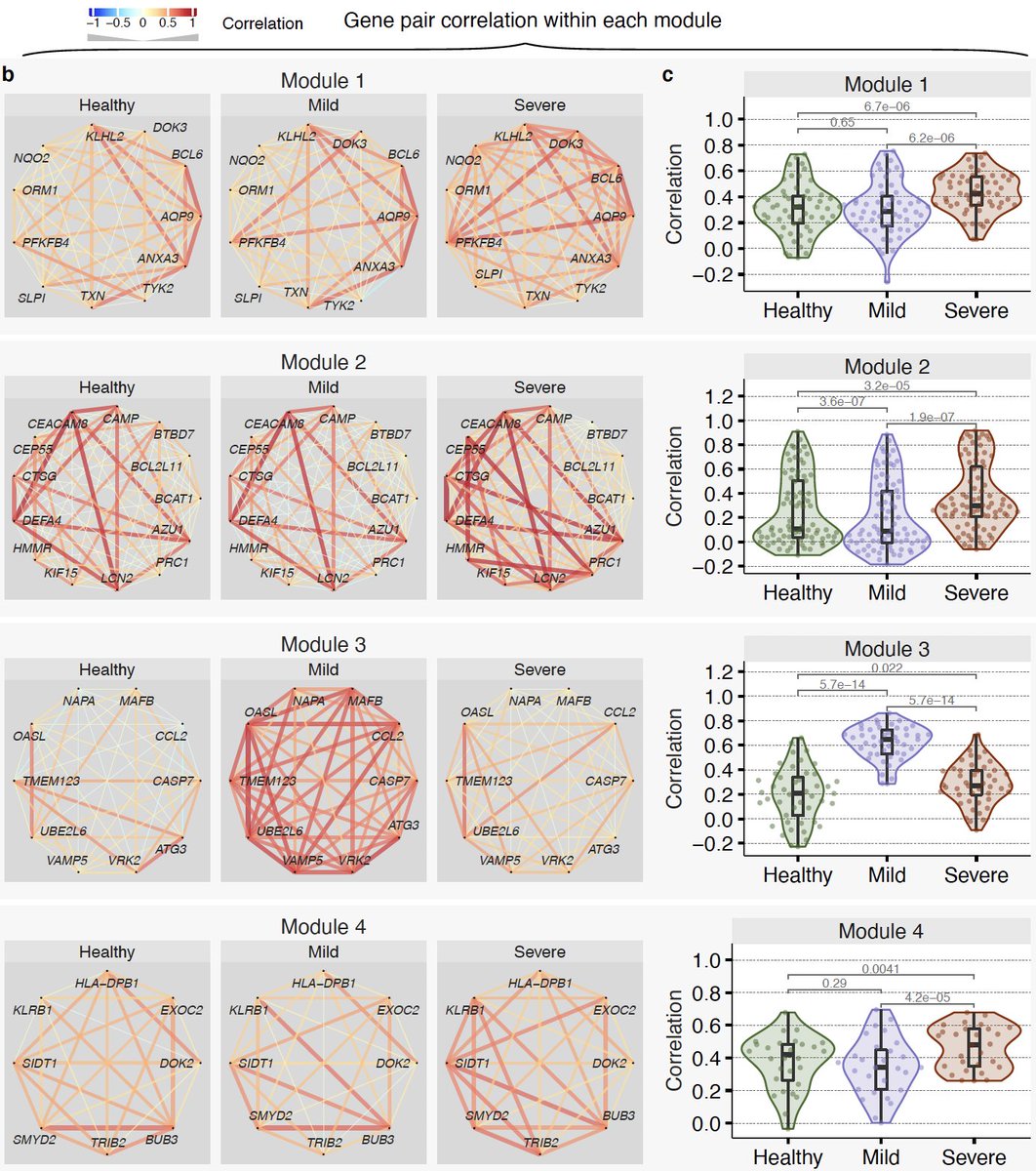
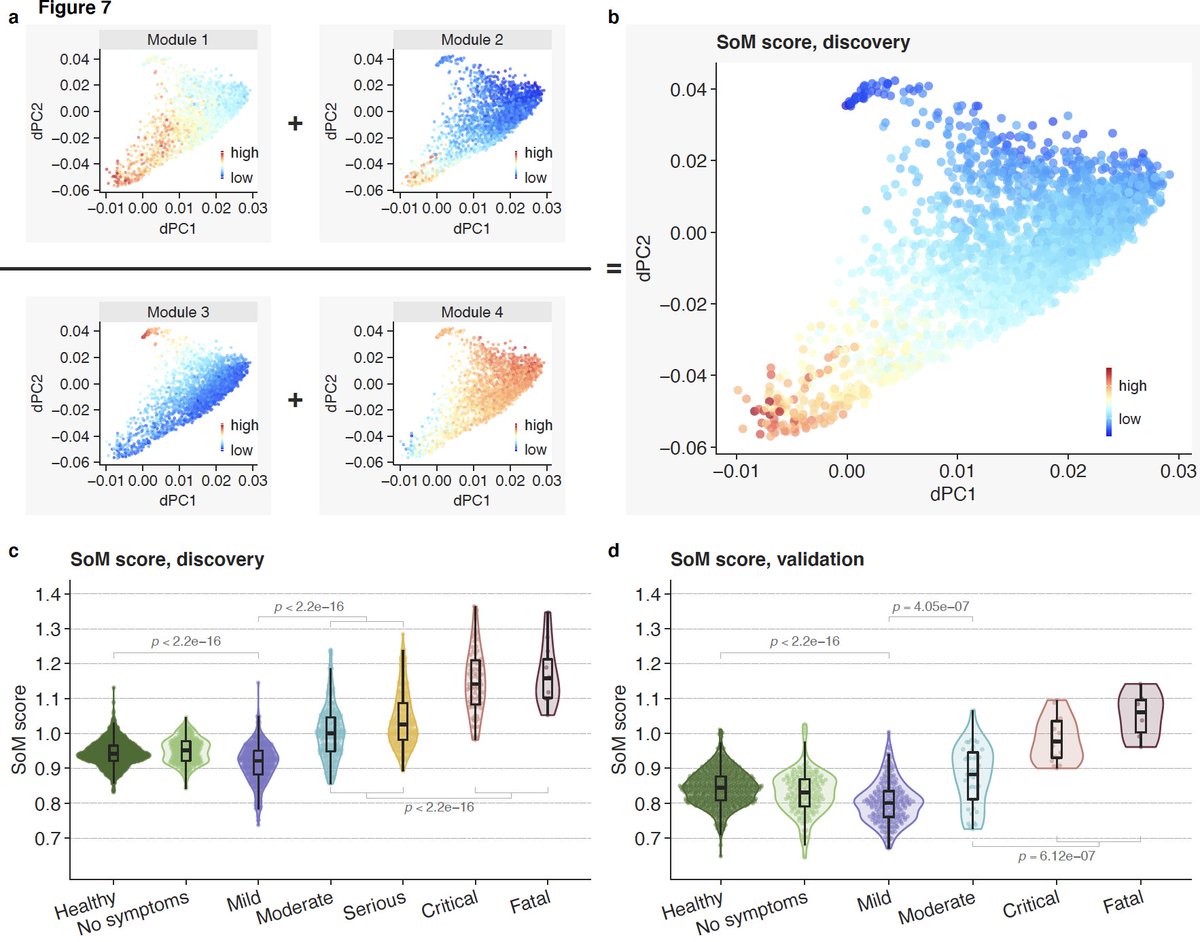
14/ We find these genes fall in 4 modules – 2 detrimental (module 1&2) and 2 protective (module 3&4). Expression of genes in detrimental modules increase with severity. Genes in protective modules higher in mild compared to severe.
15/ Interestingly though, distribution of pairwise correlation between genes in these modules also change with severity compared to healthy controls. Violin plots show distribution of pairwise correlations.
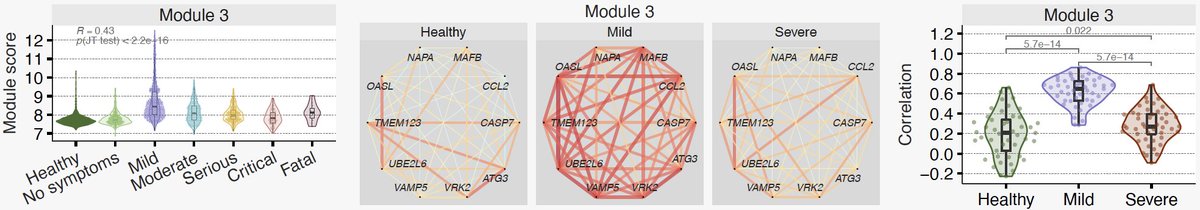
16/ Module 3 is our favorite. It is higher in those with mild outcomes, but barely changes in those with severe outcomes compared to healthy controls.
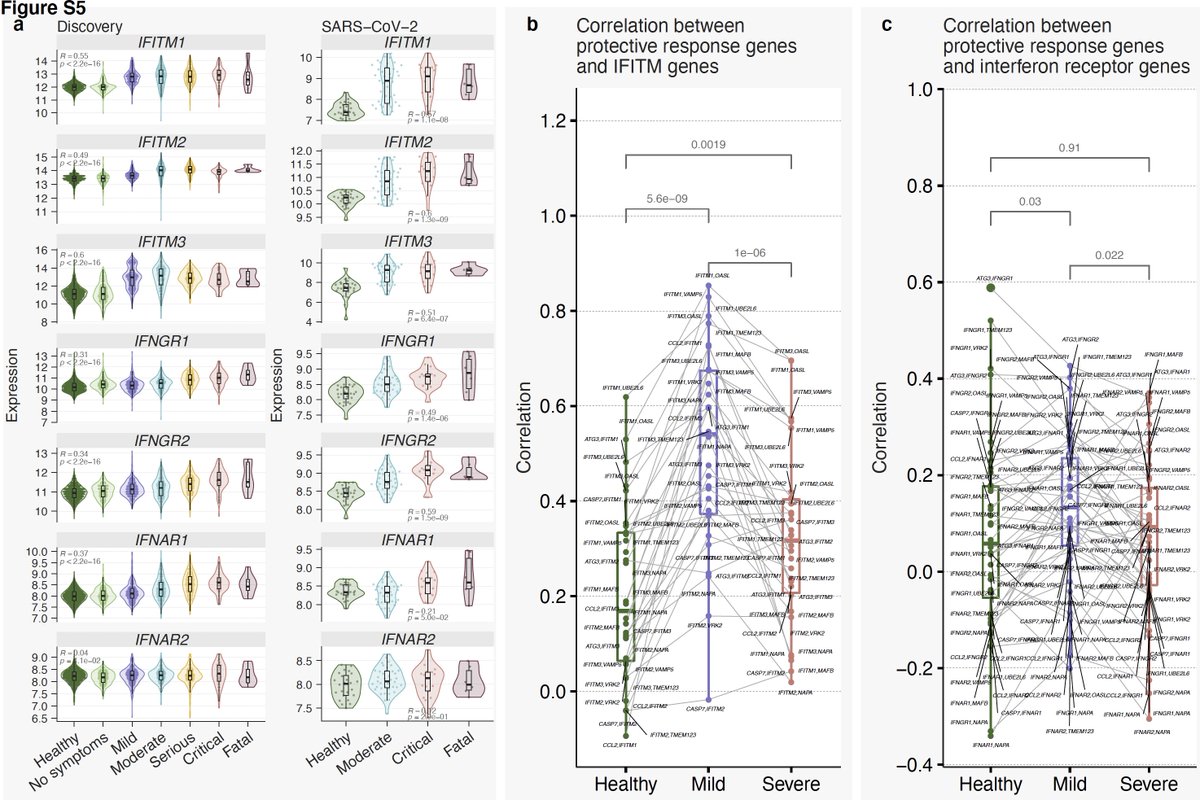
17/ Strikingly, we see decoupling between the protective response and IFN response in severe patients. Notice correlation between genes in module 3 and type I and II IFN receptors and IFITMs is much higher in patients with mild outcome than those with severe outcome.
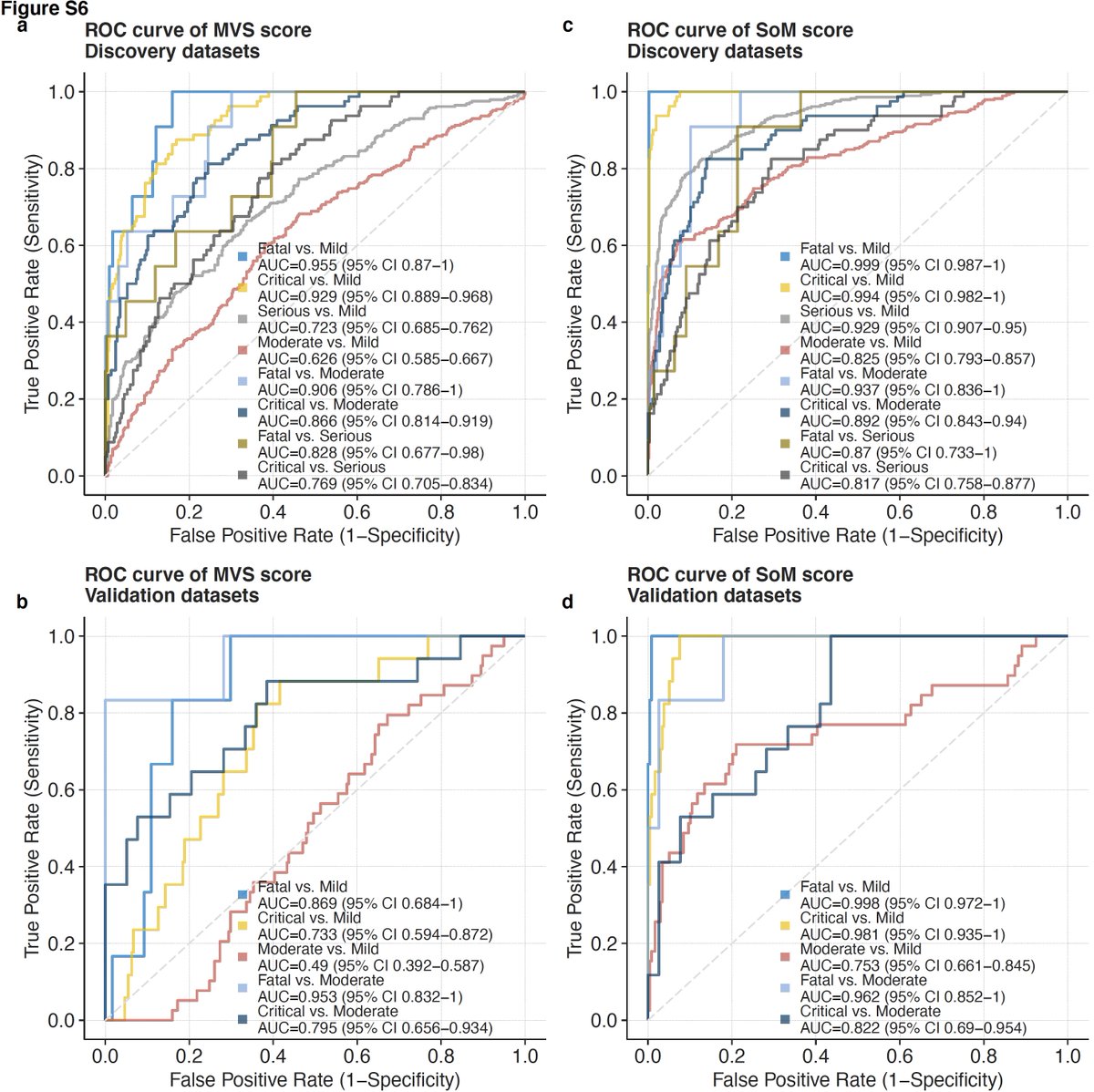
19/ SoM score distinguishes mild and severe viral infections with higher accuracy than the MVS score.
20/ Summary below and more details in the manuscript https://medrxiv.org/cgi/content/short/2020.10.02.20205880v1.">https://medrxiv.org/cgi/conte... So what do these results mean for the current pandemic and the ones that will invariably come in the future?
21/ Our results show that host response in patients with severe COVID-19 is highly similar to those with severe outcome with other viruses. That& #39;s a blessing in disguise because it presents a unique opportunity to develop tools we lack in global pandemic preparedness...
22/22 Immune system has learned to recognize viruses, novel or re-emerging and how to respond to them. We can leverage what it learned over millions of years of evolution as a general response to viruses and develop better diagnostics and prognostics, and host-directed therapies.

 Read on Twitter
Read on Twitter![[Thread] 1/22 Check out our pan-virus analysis! Tremendous teamwork by @bitsoftime @adityarao95 Denis Dermadi, @jiaying_toh, @ladoctorajones @mdonato1138 and @yemlooo with our collaborators @LabHeath @isbsci and @yehudithasin and Yudong He @inflammatix_inc https://medrxiv.org/cgi/conte... [Thread] 1/22 Check out our pan-virus analysis! Tremendous teamwork by @bitsoftime @adityarao95 Denis Dermadi, @jiaying_toh, @ladoctorajones @mdonato1138 and @yemlooo with our collaborators @LabHeath @isbsci and @yehudithasin and Yudong He @inflammatix_inc https://medrxiv.org/cgi/conte...](https://pbs.twimg.com/media/EjmL1_cVoAAvd7R.jpg)