1/ Me and @jaralaus made a tutorial on how to perform sequential bayes factor testing (using #rstats) to stop a study early in research fields were samples are small and comes with large monetary and ethical costs. https://twitter.com/medrxivpreprint/status/1306989332994101253?s=20">https://twitter.com/medrxivpr... Below, a thread:
2/ The tutorial are primarily inspired by the excellent paper by @nicebread303, @EJWagenmakers and more: https://osf.io/kf5q9/ .">https://osf.io/kf5q9/&qu... However, our target audience is the field of molecular imaging research where BF hypothesis testing is rare (<- understatement).
3/ We therefore put a lot of focus on explaining the basics, mostly use conventional/default settings, look at two-sample and a paired design, and generally try to make it relevant for a molecular imaging setting.
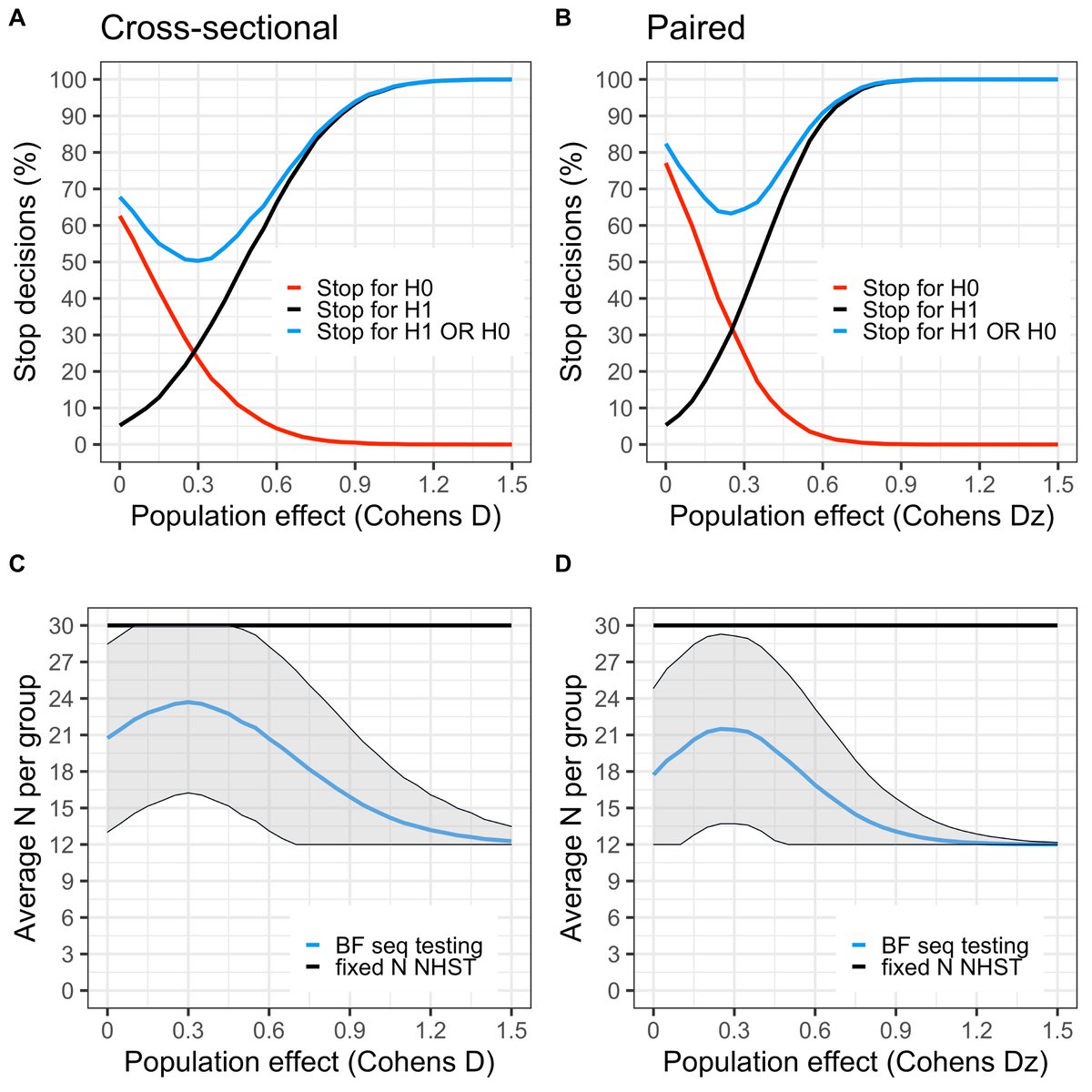
4/ We start with slowly boiling the frog, showing how to stop only in favour of the alternative hypothesis, and benchmark everything against the one and only way inference is made in the field today: the classical fixed N NHST.
5/ We then turn up the heat and introduce stopping for H0 as well. For small samples it can be difficult to obtain support for H0 with a two-tailed "default" H1. We instead make a strong recommendation of going one-tailed, based partly on this thread: https://twitter.com/ppsigray/status/1219698711212806144?s=20">https://twitter.com/ppsigray/...
6/ By introducing early stop in favor of H0, our hope is that more researchers will be able to abort expensive studies of null or near null-effects. Thus, reducing temptations to commit QRPs (hopefully…), and allowing for reallocation of resources.
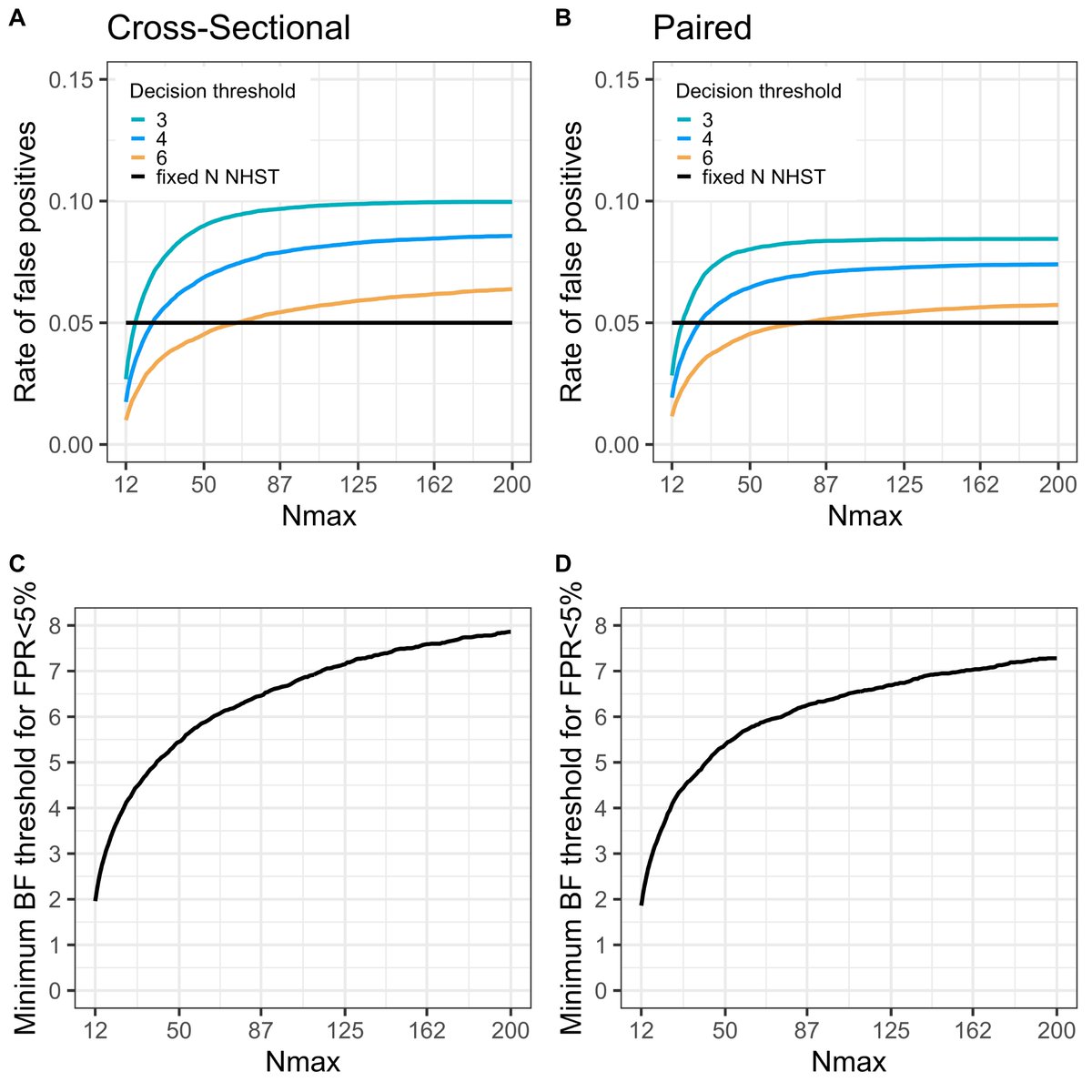
7/ In our field, we will likely always have to pick a fixed max N, even for a seq-design, due to ethical and radiation safety issues. This is interesting, because when you set a maximum N, it’s easy to pick settings that allow for *exact* control over false positive errors.
(8/ Well I say “exact”, but the settings are still estimated from simulations. If anybody knows of an analytical approximation/solution to this, I’ve would be more than happy for pointers because we have not been able to find any.)
9/ E.g., with a max possible N=30/group (start seq-testing at N=12/group), a stop threshold of 4.25 will keep the false positive rate at 5%. With a higher N-max, the BF threshold needs to increase (and eventually reach an asymptote); and with a lower N-max, it decreases.
10/ Why 5%? Well, it is what it is: I have yet seen a non-SPM paper in clinical molecular imaging that used a different threshold for false positives… But the code can be modified to find the threshold for whatever false positive rate your heart desires. https://github.com/pontusps/Early_stopping_in_PET">https://github.com/pontusps/...
11/ Depending on the true effect size, you will lose some “power” (not more than 10%) using this procedure, compared to fixed N NHST, but on the other hand you are allowed to peak at data after the inclusion of e.g. each case-control pair and can save some resources!
12/ We show this by applying what we learned to a real positron emission tomography dataset and see that we could have stopped at 27 subj/group instead of the full 40/group. Since it’s not unusual for single research PET scan to cost around 50k Euro, that’s a lot of money…
13/ Perhaps weird to discuss “power/error control” when it comes to BF, and many would likely think this is a silly use of the metric. But we think it makes sense when planning to perform a study with seq BF, and it’s a nice gateway for a field that only heard about NHST.
14/14 We would love to get some feedback, @EJWagenmakers, @nicebread303, @JeffRouder, @richarddmorey, given time and interest of-course. Maybe also of interest to @TurkuPETCentre, @dsquintana?

 Read on Twitter
Read on Twitter