1/24 - Can we decipher order from cancer genomes? Our new paper “Pervasive chromosomal instability and karyotype order in tumour evolution” comes out in Nature today: https://www.nature.com/articles/s41586-020-2698-6">https://www.nature.com/articles/... @BCRFcure @TheCrick @CRUKresearch @RosetreesT @uclcancer @SwantonLab @royalsociety
2/24 - Chromosomal instability (CIN) is common in cancer and consists of dynamic changes in chromosome number and structure. This instability can result in somatic copy number alterations - SCNAs - which may provide a substrate for tumour evolution.
3/24 - In our study, we aim to better understand the SCNA landscape in cancer. How prevalent are SCNAs in different tumour types? How homogeneous (clonal) or heterogeneous (subclonal) are SCNAs in tumours? Is CIN an ongoing process? Does the SCNA landscape change with metastasis?
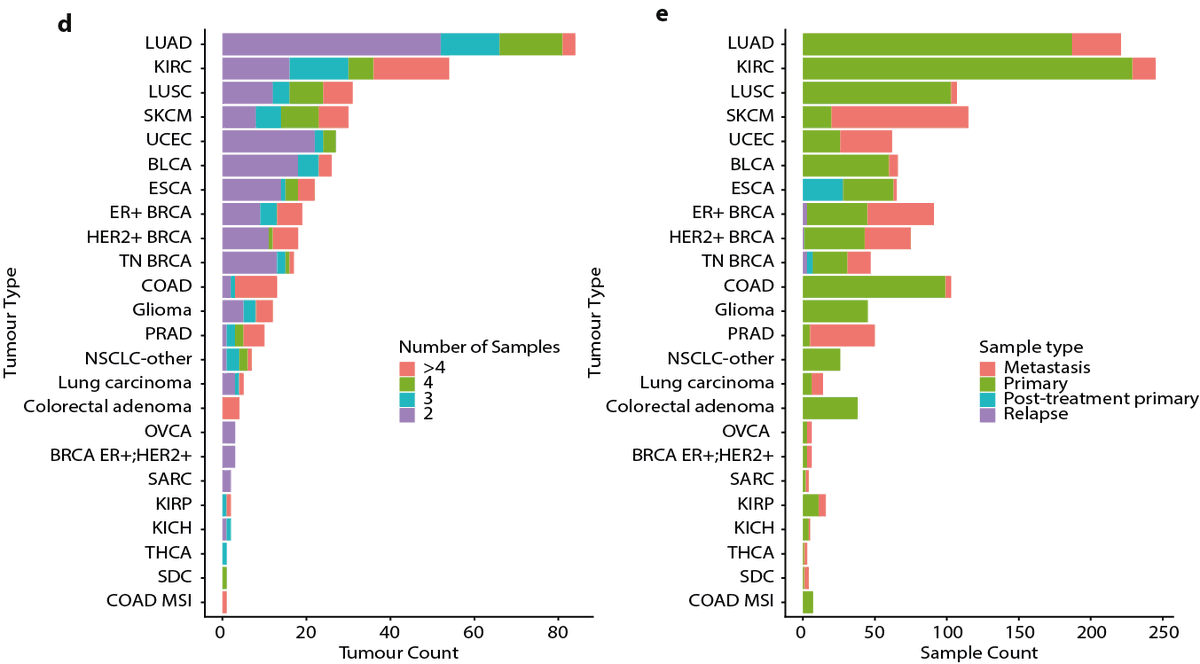
4/24 - To explore these questions, @tbkwatkins, @LimEmilia with the @rfschwarz and @NickyMcGranahan labs gathered sequencing data for 1421 samples from 394 tumours, representing 22 tumour types, with 152 tumours having at least one primary and at least one metastatic sample.
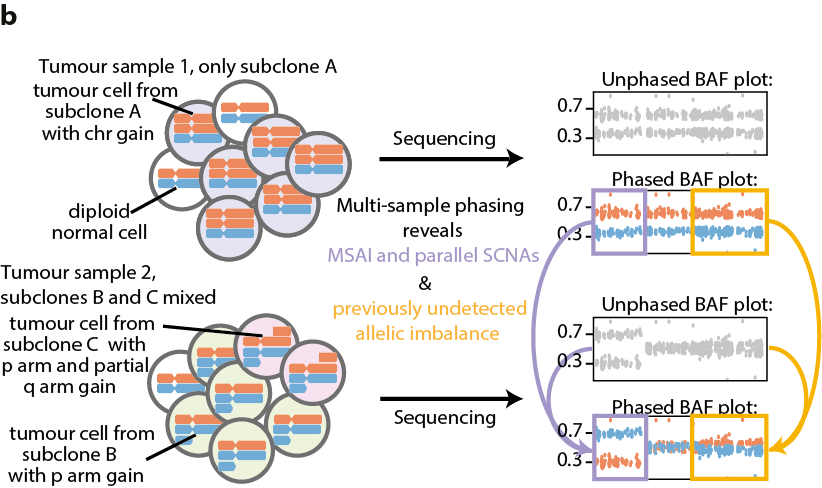
5/24 - To quantify allele-specific SCNA heterogeneity in tumour samples we identified gains, losses, loss of heterozygosity, and used multi-sample phasing to identify mirrored subclonal allelic imbalance (MSAI), parallel evolution and previously undetected allelic imbalance.
6/24 - MSAI is caused by SCNAs that disrupt the same genomic region but affect different parental alleles in separate tumour subclones and cannot be observed from a single bulk sample. When MSAI derives from similar SCNAs (e.g. independent gains) it indicates parallel evolution.
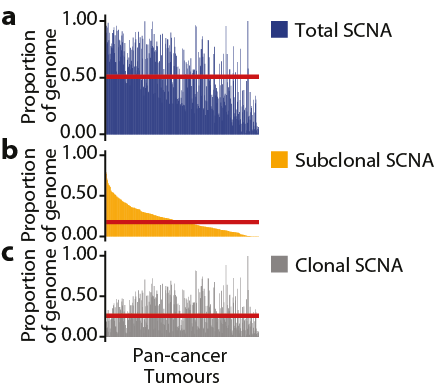
7/24 - Clonal (grey) and subclonal (orange) SCNAs were observed across the genome. A median of 26% of the genome was subject to clonal SCNAs and 18% to subclonal. 45% of tumours harboured subclonal SCNAs in >20% of the genome suggesting that CIN is ongoing and pervasive.
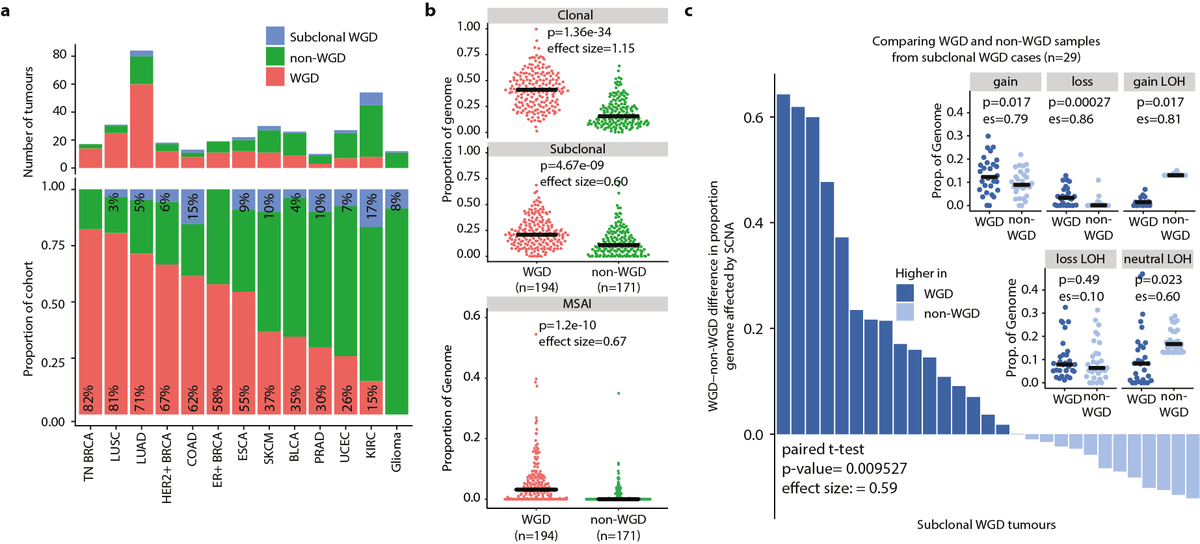
8/24 - The timing of SCNAs varied across tumour types. For instance, we found that despite a comparable total proportion of the genome affected by SCNAs between lung adenocarcinoma and HER2+ breast cancer, in lung adenocarcinoma a larger proportion of SCNAs were subclonal.
9/24 - We found that 49% of tumours showed clonal whole-genome doubling (WGD) and 7% subclonal WGD. Interestingly, WGD was linked to increased incidence of clonal and subclonal SCNA as well as MSAI, suggesting that WGD was often an early transformative event in tumour evolution.
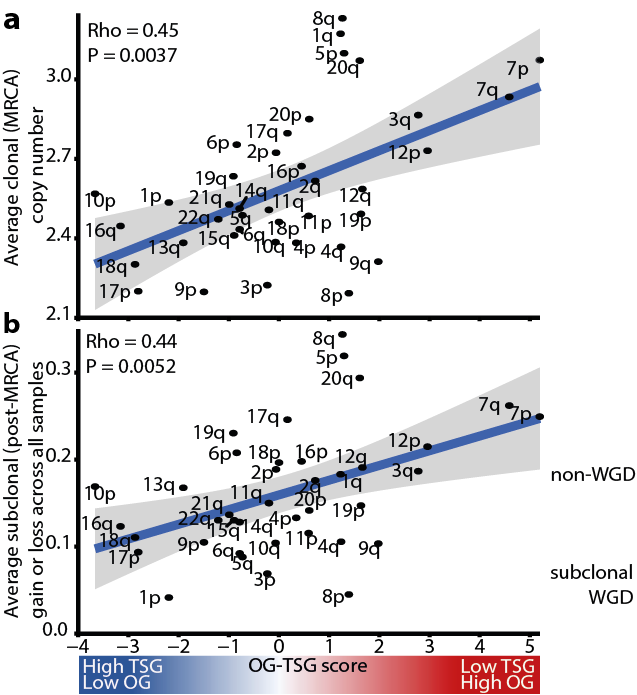
10/24 – Next, to investigate the degree to which SCNAs are shaped by neutral evolution or selection, we used a Markov chain model to analyse whether the propensity for a chromosome arm to be gained or lost was related to the density of tumour-suppressor genes and oncogenes.
11/24 - @marpetkov inferred the SCNA landscape of the most recent common ancestor (MRCA) for each tumour. Consistent with ongoing selection, the OG–TSG score significantly correlated with the burden of arm-level alterations in the MRCA and with subclonal arm-level alterations.
12/24 - Markov chain modelling by @sergi_elizalde and @Samuel_Bakhoum using the chromosome arm OG-TSG scores best predicted the profile of subclonal SCNAs in 59% of the cohort but and especially in WGD cases, further suggesting that WGD may support subsequent genome remodelling.
13/24 - However, in 41% of our cohort the neutral (no OG-TSG scores) or scrambled (permuted OG-TSG scores) models best predicted the subclonal SCNAs, potentially reflecting the evolution of a neutral karyotype or the need for tumour-type-specific chromosome arm score weightings.
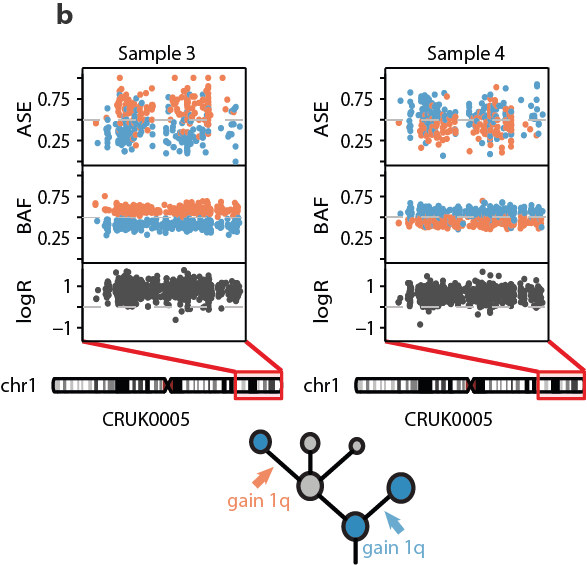
14/24 - Parallel evolution of SCNAs revealed through MSAI detection, identifying similar events within distinct subclones in the same tumour, was observed in 37% of tumours. Allele-specific expression tracked parallel evolutionary events that originated from distinct haplotypes.
15/24 - The most prominent parallel gains included those overlapping chromosomes 1q21.3–q44, which encompasses BCL9, MCL1, and ARNT/HIF1B, 5p15.33 which includes TERT, and 8q24.1, which encompasses MYC.
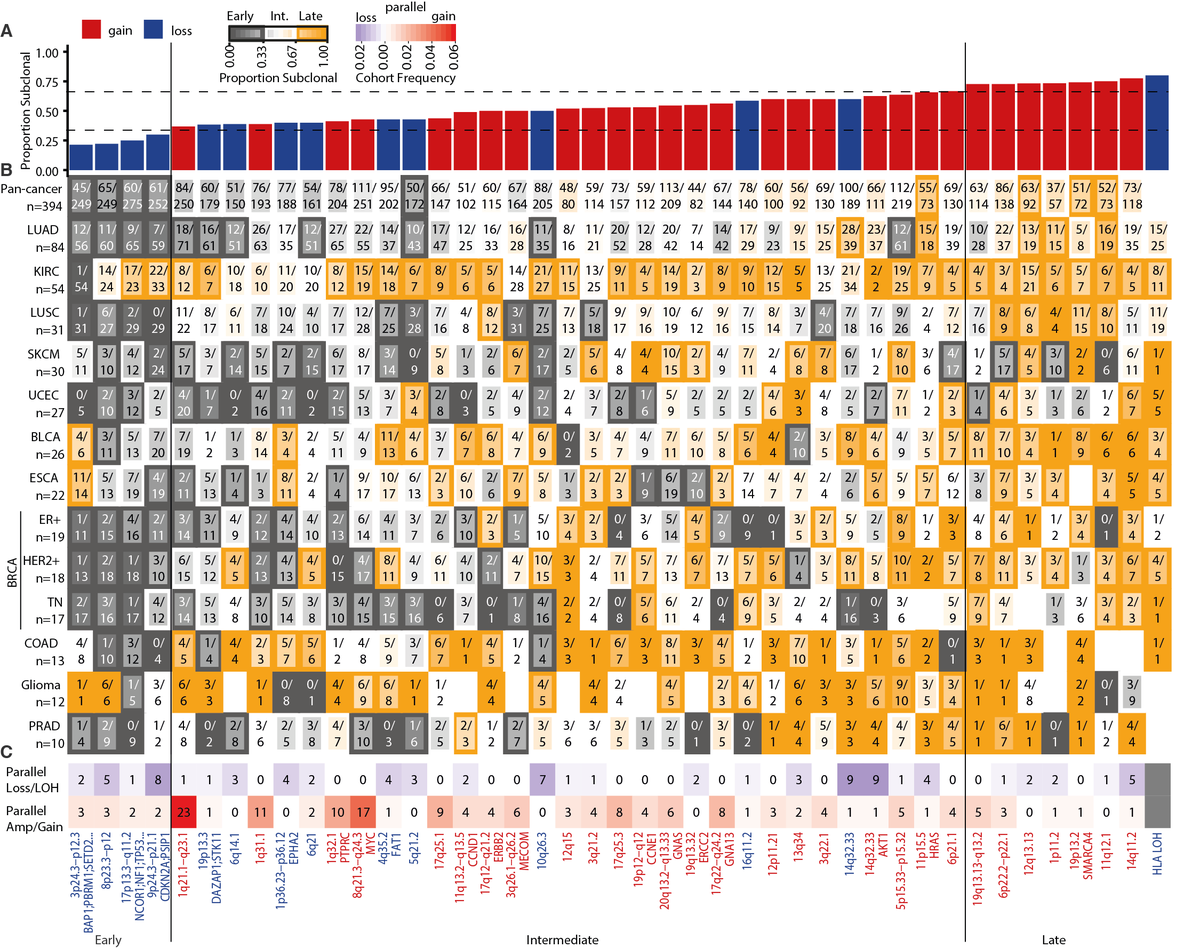
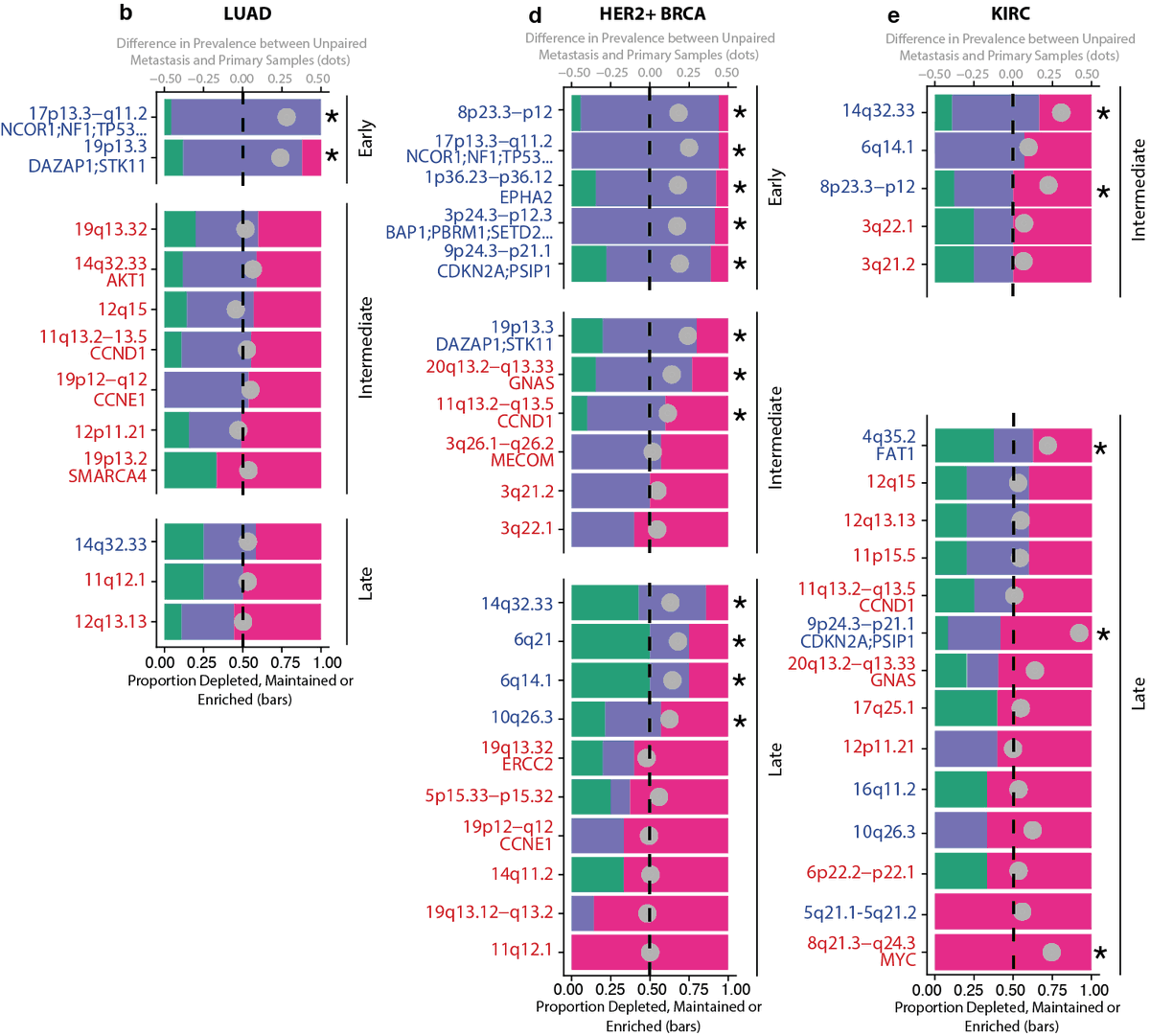
16/24 - We identified consensus GISTIC2.0 peak regions across tumour types and categorised their timing: early (potentially implicated in tumourigenesis), intermediate/late (potentially involved in tumour maintenance and progression) and mapped parallel evolution events to them.
17/24 - Loss peaks were more likely to be early and enriched in TSGs compared to gain peaks, which were enriched in known oncogenes. For example, 9p24.3-p21.1 (encompassing CDK2NA and PSIP1) loss was early in 7 of 13 tumour types.
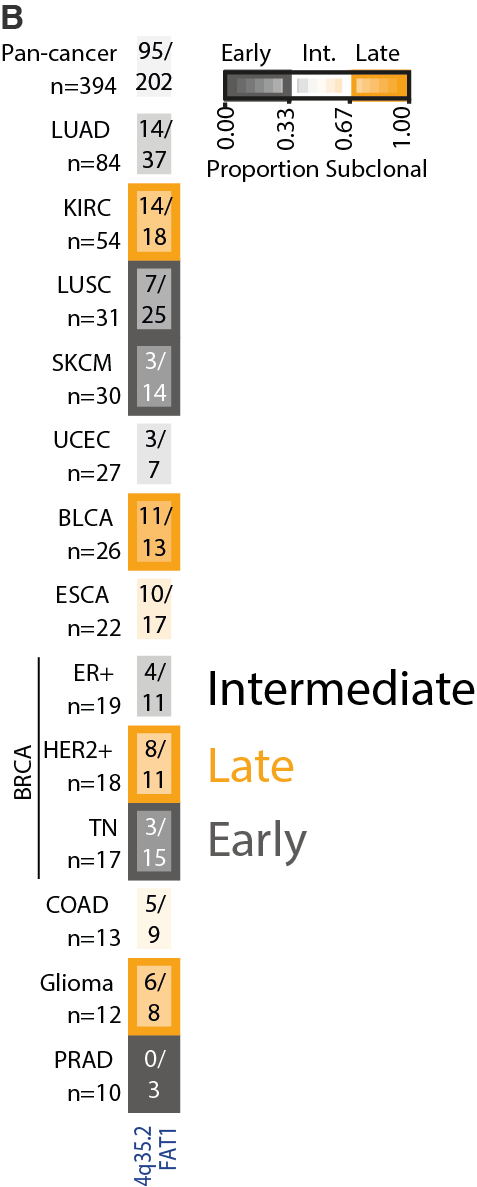
18/24 - Intriguingly, the timing of other peak regions varied by tumour type. For example, the loss peak at chromosome 4q35.2 encompassing the tumour suppressor FAT1 was early in triple-negative breast cancer, intermediate in ER+ breast cancer and late in HER2+ breast cancer.
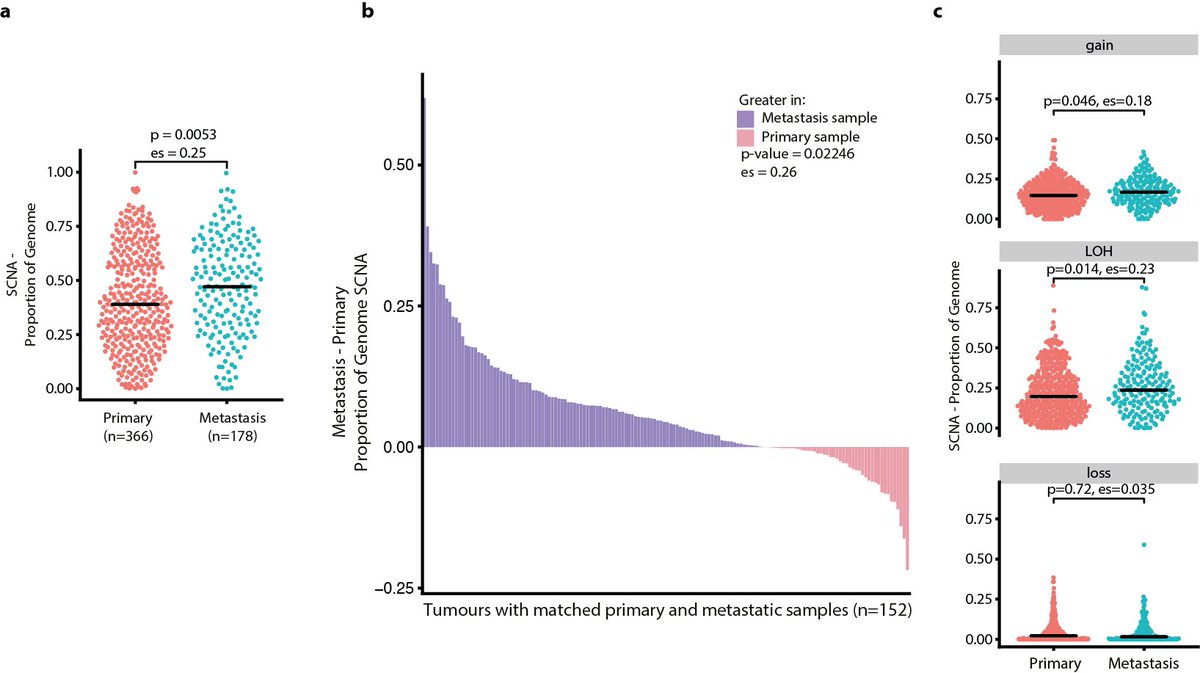
19/24 - Finally, we explored how the SCNA landscape alters with metastasis. We found a higher proportion of the genome affected by SCNAs in metastases compared with primary tumours. SCNAs were more frequently clonal in metastases with LOH events showing the greatest increase.
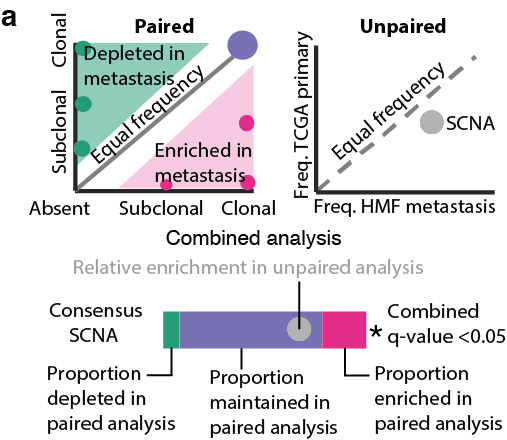
20/24 - We investigated our timed consensus SCNAs in metastasis performing a combined analysis using both paired samples from our cohort and 2631 unpaired metastatic samples from the TCGA and 1024 Hartwig Medical Foundation.
21/24 - Amongst other SCNAs, 19p13.3 (STK11) loss, early in lung adenocarcinoma, 11q13.2-q13.5 (CCND1) gain, intermediate in HER2+ breast cancer, gain of 8q21.3–q24.3 (MYC), late in clear cell renal carcinoma were enriched in metastases.
22/24 - In conclusion, clonal and subclonal SCNAs are pervasive across tumour types and tend to occur as ordered events which may reflect continuous optimisation of the fitness landscape. Our work highlights the importance of ongoing CIN during tumour evolution and metastasis.
23/24 - We analysed multiple bulk samples from each tumour. However, allele-specific single-cell SCNA calling is needed to fully determine the extent and importance of subclonal SCNAs in tumour evolution. @ZaccaSimo and @benjraphael’s CHISEL is an exciting step in this direction.
24/24 - Thank you to collaborators @SwantonLab @rfschwarz @NickyMcGranahan @Samuel_Bakhoum @marpetkov @sergi_elizalde and many others listed on the paper, without whom this work would not have been possible.

 Read on Twitter
Read on Twitter