Our #research into #pandemic multidrug-resistant E. coli O25b:H4:ST131 is FINALLY out in @FrontMicrobiol https://abs.twimg.com/emoji/v2/... draggable="false" alt="‼️" title="Doppeltes Ausrufezeichen" aria-label="Emoji: Doppeltes Ausrufezeichen"> Using WGS to show companion animals represent occasional spillover hosts rather than primary reservoirs for the ST131 lineage...a thread
https://abs.twimg.com/emoji/v2/... draggable="false" alt="‼️" title="Doppeltes Ausrufezeichen" aria-label="Emoji: Doppeltes Ausrufezeichen"> Using WGS to show companion animals represent occasional spillover hosts rather than primary reservoirs for the ST131 lineage...a thread https://abs.twimg.com/emoji/v2/... draggable="false" alt="⬇️" title="Pfeil nach unten" aria-label="Emoji: Pfeil nach unten">1/7
https://abs.twimg.com/emoji/v2/... draggable="false" alt="⬇️" title="Pfeil nach unten" aria-label="Emoji: Pfeil nach unten">1/7
Paper https://abs.twimg.com/emoji/v2/... draggable="false" alt="👉" title="Rückhand Zeigefinger nach rechts" aria-label="Emoji: Rückhand Zeigefinger nach rechts"> https://www.frontiersin.org/articles/10.3389/fmicb.2020.01968/full">https://www.frontiersin.org/articles/...
https://abs.twimg.com/emoji/v2/... draggable="false" alt="👉" title="Rückhand Zeigefinger nach rechts" aria-label="Emoji: Rückhand Zeigefinger nach rechts"> https://www.frontiersin.org/articles/10.3389/fmicb.2020.01968/full">https://www.frontiersin.org/articles/...
Paper
Recently, #ST131 has emerged & spread globally. Dissemination is unclear in #Australia. There’re reports of commonality between ST131 from humans & pets. Here, using #WGS, we investigated pet-sourced E. coli on an Australia-wide level against a global collection  https://abs.twimg.com/emoji/v2/... draggable="false" alt="🇦🇺" title="Flagge von Australien" aria-label="Emoji: Flagge von Australien">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🇦🇺" title="Flagge von Australien" aria-label="Emoji: Flagge von Australien"> https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌏" title="Asien-Australien auf dem Globus" aria-label="Emoji: Asien-Australien auf dem Globus"> 2/7
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌏" title="Asien-Australien auf dem Globus" aria-label="Emoji: Asien-Australien auf dem Globus"> 2/7
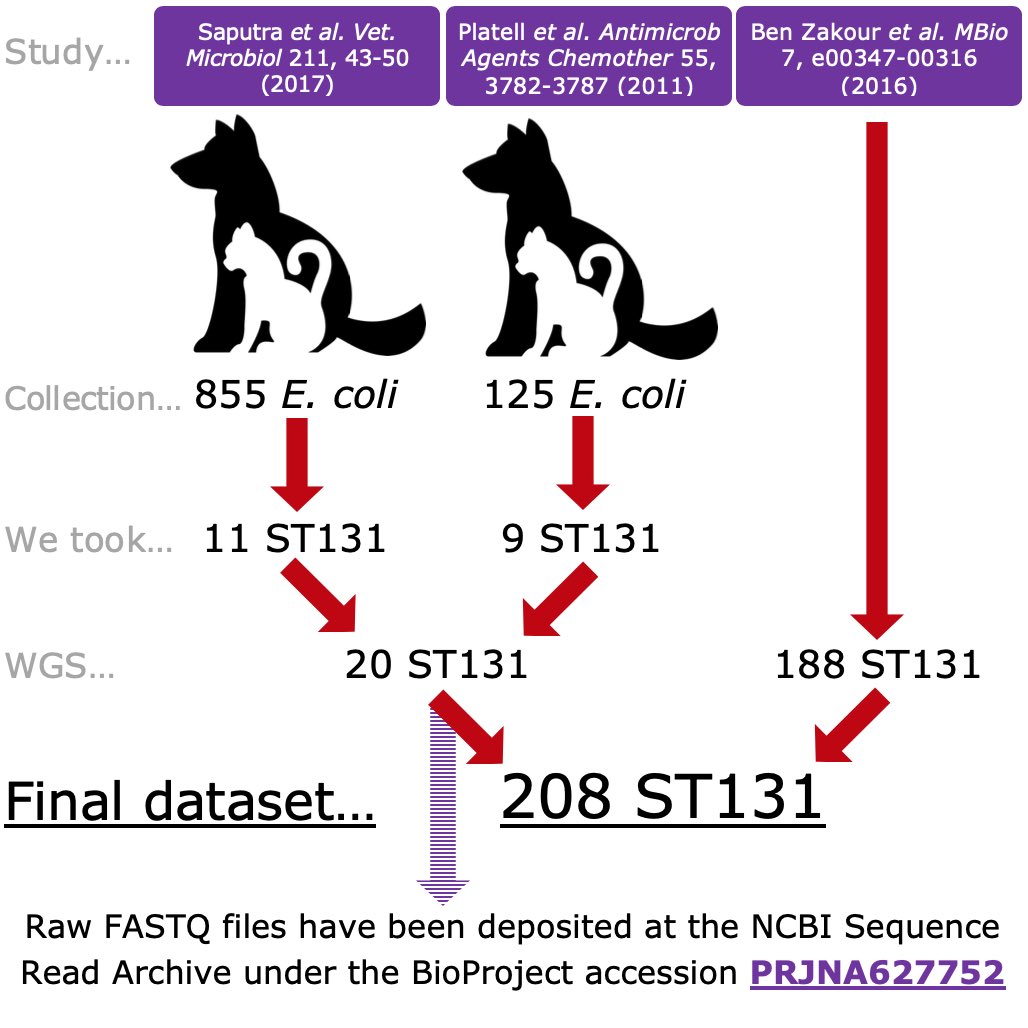
We used  https://abs.twimg.com/emoji/v2/... draggable="false" alt="3️⃣" title="Tastenkappe Ziffer 3" aria-label="Emoji: Tastenkappe Ziffer 3"> sources of isolates & compared characteristics of FQR E. coli from humans & pets, using PCR-based phylotyping & WGS. We include #genomic data from Aussie isolates over 2-years & a previous study by @genomiss in @mbiojournal
https://abs.twimg.com/emoji/v2/... draggable="false" alt="3️⃣" title="Tastenkappe Ziffer 3" aria-label="Emoji: Tastenkappe Ziffer 3"> sources of isolates & compared characteristics of FQR E. coli from humans & pets, using PCR-based phylotyping & WGS. We include #genomic data from Aussie isolates over 2-years & a previous study by @genomiss in @mbiojournal  https://abs.twimg.com/emoji/v2/... draggable="false" alt="👉" title="Rückhand Zeigefinger nach rechts" aria-label="Emoji: Rückhand Zeigefinger nach rechts"> https://mbio.asm.org/content/7/2/e00347-16.abstract">https://mbio.asm.org/content/7... [110 citations] 3/7
https://abs.twimg.com/emoji/v2/... draggable="false" alt="👉" title="Rückhand Zeigefinger nach rechts" aria-label="Emoji: Rückhand Zeigefinger nach rechts"> https://mbio.asm.org/content/7/2/e00347-16.abstract">https://mbio.asm.org/content/7... [110 citations] 3/7
We show the epidemiological associations of these isolates is leading to the clinically relevant conclusion that the transfer of #AMR E. coli ST131 between humans & pets is much less frequent than assumed 4/7
This work showcases skills from a great collaborative team, incl.
 https://abs.twimg.com/emoji/v2/... draggable="false" alt="💊" title="Tablette" aria-label="Emoji: Tablette">Amanda Kidsley & Darren Trott ( @UniofAdelaide)
https://abs.twimg.com/emoji/v2/... draggable="false" alt="💊" title="Tablette" aria-label="Emoji: Tablette">Amanda Kidsley & Darren Trott ( @UniofAdelaide)
 https://abs.twimg.com/emoji/v2/... draggable="false" alt="💊" title="Tablette" aria-label="Emoji: Tablette">the Australian Group for #AMR affiliated w/ @AusAntibiotics
https://abs.twimg.com/emoji/v2/... draggable="false" alt="💊" title="Tablette" aria-label="Emoji: Tablette">the Australian Group for #AMR affiliated w/ @AusAntibiotics
 https://abs.twimg.com/emoji/v2/... draggable="false" alt="💊" title="Tablette" aria-label="Emoji: Tablette"> @beatsonlab & @MarkSchembri7
https://abs.twimg.com/emoji/v2/... draggable="false" alt="💊" title="Tablette" aria-label="Emoji: Tablette"> @beatsonlab & @MarkSchembri7
 https://abs.twimg.com/emoji/v2/... draggable="false" alt="💊" title="Tablette" aria-label="Emoji: Tablette"> @mrsmolly01
https://abs.twimg.com/emoji/v2/... draggable="false" alt="💊" title="Tablette" aria-label="Emoji: Tablette"> @mrsmolly01
 https://abs.twimg.com/emoji/v2/... draggable="false" alt="💊" title="Tablette" aria-label="Emoji: Tablette"> @sam_manna3
https://abs.twimg.com/emoji/v2/... draggable="false" alt="💊" title="Tablette" aria-label="Emoji: Tablette"> @sam_manna3
 https://abs.twimg.com/emoji/v2/... draggable="false" alt="👉" title="Rückhand Zeigefinger nach rechts" aria-label="Emoji: Rückhand Zeigefinger nach rechts">& many more I can’t find on twitter
https://abs.twimg.com/emoji/v2/... draggable="false" alt="👉" title="Rückhand Zeigefinger nach rechts" aria-label="Emoji: Rückhand Zeigefinger nach rechts">& many more I can’t find on twitter https://abs.twimg.com/emoji/v2/... draggable="false" alt="‼️" title="Doppeltes Ausrufezeichen" aria-label="Emoji: Doppeltes Ausrufezeichen"> 5/7
https://abs.twimg.com/emoji/v2/... draggable="false" alt="‼️" title="Doppeltes Ausrufezeichen" aria-label="Emoji: Doppeltes Ausrufezeichen"> 5/7
This project was funded by an @arc_gov_au grant w/ @Zoetis Australia as a main industry partner. This work was also supported in part by the @DeptVetAffairs. We acknowledge the assistance of the private, government & university veterinary diagnostic laboratories in Australia 6/7
Sequence read data has been submitted to the @NCBI Sequence Read Archive (SRA) under BioProject  https://abs.twimg.com/emoji/v2/... draggable="false" alt="👉" title="Rückhand Zeigefinger nach rechts" aria-label="Emoji: Rückhand Zeigefinger nach rechts">PRJNA627752
https://abs.twimg.com/emoji/v2/... draggable="false" alt="👉" title="Rückhand Zeigefinger nach rechts" aria-label="Emoji: Rückhand Zeigefinger nach rechts">PRJNA627752 https://abs.twimg.com/emoji/v2/... draggable="false" alt="👈" title="Rückhand Zeigefinger nach links" aria-label="Emoji: Rückhand Zeigefinger nach links">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="👈" title="Rückhand Zeigefinger nach links" aria-label="Emoji: Rückhand Zeigefinger nach links">
Sequence data https://abs.twimg.com/emoji/v2/... draggable="false" alt="👉" title="Rückhand Zeigefinger nach rechts" aria-label="Emoji: Rückhand Zeigefinger nach rechts"> https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA627752
https://abs.twimg.com/emoji/v2/... draggable="false" alt="👉" title="Rückhand Zeigefinger nach rechts" aria-label="Emoji: Rückhand Zeigefinger nach rechts"> https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA627752
de">https://www.ncbi.nlm.nih.gov/bioprojec... novo assemblies https://abs.twimg.com/emoji/v2/... draggable="false" alt="👉" title="Rückhand Zeigefinger nach rechts" aria-label="Emoji: Rückhand Zeigefinger nach rechts"> https://github.com/RhysWhite/ST131_19_Pets_AU
https://abs.twimg.com/emoji/v2/... draggable="false" alt="👉" title="Rückhand Zeigefinger nach rechts" aria-label="Emoji: Rückhand Zeigefinger nach rechts"> https://github.com/RhysWhite/ST131_19_Pets_AU
7/7">https://github.com/RhysWhite...
Sequence data
de">https://www.ncbi.nlm.nih.gov/bioprojec... novo assemblies
7/7">https://github.com/RhysWhite...

 Read on Twitter
Read on Twitter Using WGS to show companion animals represent occasional spillover hosts rather than primary reservoirs for the ST131 lineage...a threadhttps://abs.twimg.com/emoji/v2/... draggable="false" alt="⬇️" title="Pfeil nach unten" aria-label="Emoji: Pfeil nach unten">1/7Paperhttps://abs.twimg.com/emoji/v2/... draggable="false" alt="👉" title="Rückhand Zeigefinger nach rechts" aria-label="Emoji: Rückhand Zeigefinger nach rechts"> https://www.frontiersin.org/articles/..." title="Our #research into #pandemic multidrug-resistant E. coli O25b:H4:ST131 is FINALLY out in @FrontMicrobiolhttps://abs.twimg.com/emoji/v2/... draggable="false" alt="‼️" title="Doppeltes Ausrufezeichen" aria-label="Emoji: Doppeltes Ausrufezeichen"> Using WGS to show companion animals represent occasional spillover hosts rather than primary reservoirs for the ST131 lineage...a threadhttps://abs.twimg.com/emoji/v2/... draggable="false" alt="⬇️" title="Pfeil nach unten" aria-label="Emoji: Pfeil nach unten">1/7Paperhttps://abs.twimg.com/emoji/v2/... draggable="false" alt="👉" title="Rückhand Zeigefinger nach rechts" aria-label="Emoji: Rückhand Zeigefinger nach rechts"> https://www.frontiersin.org/articles/..." class="img-responsive" style="max-width:100%;"/>
Using WGS to show companion animals represent occasional spillover hosts rather than primary reservoirs for the ST131 lineage...a threadhttps://abs.twimg.com/emoji/v2/... draggable="false" alt="⬇️" title="Pfeil nach unten" aria-label="Emoji: Pfeil nach unten">1/7Paperhttps://abs.twimg.com/emoji/v2/... draggable="false" alt="👉" title="Rückhand Zeigefinger nach rechts" aria-label="Emoji: Rückhand Zeigefinger nach rechts"> https://www.frontiersin.org/articles/..." title="Our #research into #pandemic multidrug-resistant E. coli O25b:H4:ST131 is FINALLY out in @FrontMicrobiolhttps://abs.twimg.com/emoji/v2/... draggable="false" alt="‼️" title="Doppeltes Ausrufezeichen" aria-label="Emoji: Doppeltes Ausrufezeichen"> Using WGS to show companion animals represent occasional spillover hosts rather than primary reservoirs for the ST131 lineage...a threadhttps://abs.twimg.com/emoji/v2/... draggable="false" alt="⬇️" title="Pfeil nach unten" aria-label="Emoji: Pfeil nach unten">1/7Paperhttps://abs.twimg.com/emoji/v2/... draggable="false" alt="👉" title="Rückhand Zeigefinger nach rechts" aria-label="Emoji: Rückhand Zeigefinger nach rechts"> https://www.frontiersin.org/articles/..." class="img-responsive" style="max-width:100%;"/>
 https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌏" title="Asien-Australien auf dem Globus" aria-label="Emoji: Asien-Australien auf dem Globus"> 2/7" title="Recently, #ST131 has emerged & spread globally. Dissemination is unclear in #Australia. There’re reports of commonality between ST131 from humans & pets. Here, using #WGS, we investigated pet-sourced E. coli on an Australia-wide level against a global collection https://abs.twimg.com/emoji/v2/... draggable="false" alt="🇦🇺" title="Flagge von Australien" aria-label="Emoji: Flagge von Australien">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌏" title="Asien-Australien auf dem Globus" aria-label="Emoji: Asien-Australien auf dem Globus"> 2/7" class="img-responsive" style="max-width:100%;"/>
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌏" title="Asien-Australien auf dem Globus" aria-label="Emoji: Asien-Australien auf dem Globus"> 2/7" title="Recently, #ST131 has emerged & spread globally. Dissemination is unclear in #Australia. There’re reports of commonality between ST131 from humans & pets. Here, using #WGS, we investigated pet-sourced E. coli on an Australia-wide level against a global collection https://abs.twimg.com/emoji/v2/... draggable="false" alt="🇦🇺" title="Flagge von Australien" aria-label="Emoji: Flagge von Australien">https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌏" title="Asien-Australien auf dem Globus" aria-label="Emoji: Asien-Australien auf dem Globus"> 2/7" class="img-responsive" style="max-width:100%;"/>
 sources of isolates & compared characteristics of FQR E. coli from humans & pets, using PCR-based phylotyping & WGS. We include #genomic data from Aussie isolates over 2-years & a previous study by @genomiss in @mbiojournal https://abs.twimg.com/emoji/v2/... draggable="false" alt="👉" title="Rückhand Zeigefinger nach rechts" aria-label="Emoji: Rückhand Zeigefinger nach rechts"> https://mbio.asm.org/content/7... [110 citations] 3/7" title="We used https://abs.twimg.com/emoji/v2/... draggable="false" alt="3️⃣" title="Tastenkappe Ziffer 3" aria-label="Emoji: Tastenkappe Ziffer 3"> sources of isolates & compared characteristics of FQR E. coli from humans & pets, using PCR-based phylotyping & WGS. We include #genomic data from Aussie isolates over 2-years & a previous study by @genomiss in @mbiojournal https://abs.twimg.com/emoji/v2/... draggable="false" alt="👉" title="Rückhand Zeigefinger nach rechts" aria-label="Emoji: Rückhand Zeigefinger nach rechts"> https://mbio.asm.org/content/7... [110 citations] 3/7" class="img-responsive" style="max-width:100%;"/>
sources of isolates & compared characteristics of FQR E. coli from humans & pets, using PCR-based phylotyping & WGS. We include #genomic data from Aussie isolates over 2-years & a previous study by @genomiss in @mbiojournal https://abs.twimg.com/emoji/v2/... draggable="false" alt="👉" title="Rückhand Zeigefinger nach rechts" aria-label="Emoji: Rückhand Zeigefinger nach rechts"> https://mbio.asm.org/content/7... [110 citations] 3/7" title="We used https://abs.twimg.com/emoji/v2/... draggable="false" alt="3️⃣" title="Tastenkappe Ziffer 3" aria-label="Emoji: Tastenkappe Ziffer 3"> sources of isolates & compared characteristics of FQR E. coli from humans & pets, using PCR-based phylotyping & WGS. We include #genomic data from Aussie isolates over 2-years & a previous study by @genomiss in @mbiojournal https://abs.twimg.com/emoji/v2/... draggable="false" alt="👉" title="Rückhand Zeigefinger nach rechts" aria-label="Emoji: Rückhand Zeigefinger nach rechts"> https://mbio.asm.org/content/7... [110 citations] 3/7" class="img-responsive" style="max-width:100%;"/>



