Excited to share an update on low-N protein engineering and why we think it works!
https://bit.ly/3jAV4ih ">https://bit.ly/3jAV4ih&q...
https://bit.ly/3jAV4ih ">https://bit.ly/3jAV4ih&q...
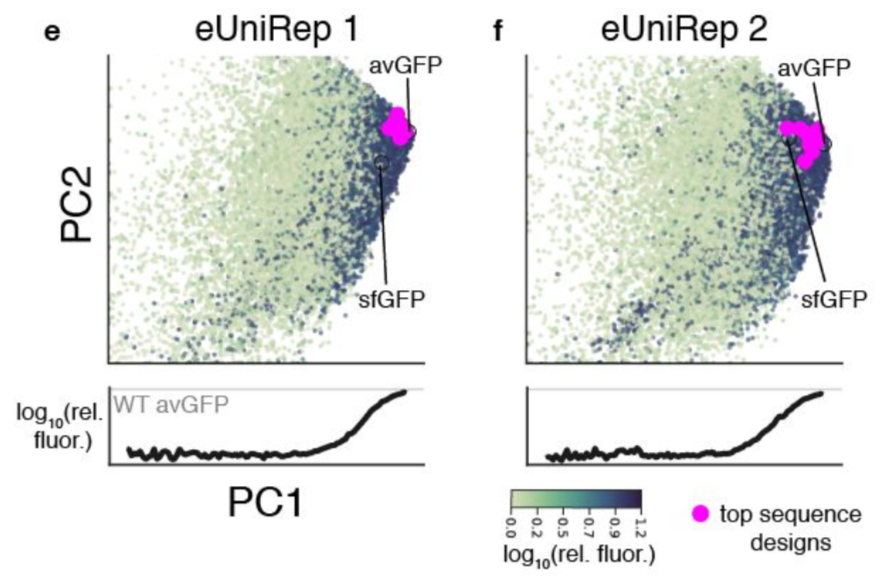
We prev showed that the first principal component (PC1) of our protein language model& #39;s representation (eUniRep) correlates with protein activity. This was interesting given the rep was learned only from raw protein sequences, without access to info about protein activity.
At a high level, this "pre-existing" knowledge means that we shouldn& #39;t need to collect as many activity measurements in order to engineer a protein. But, it& #39;s still not a full explanation for why low-N design is possible in the first place.
So we dug in some more ...
So we dug in some more ...
First we saw that the correlation between PC1 and protein activity was largely driven by PC1& #39;s ability to differentiate functional sequences from non-functional ones ...
but NOT by it& #39;s ability to differentiate sequences with >= wild-type (WT) activity. In other words, sequences with >= WT activity all just looked like WT in terms of PC1 scores.
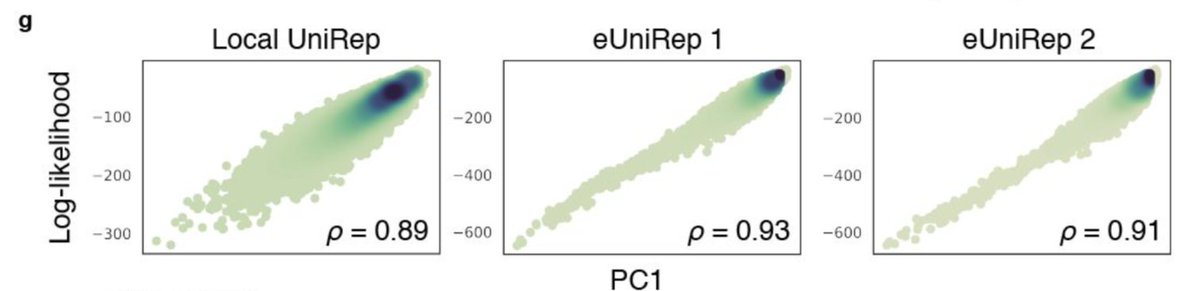
A little more digging showed that PC1 was highly correlated with sequence log-likelihood under a given UniRep model.
Because our rep learning was done using natural protein sequences, this suggests that the primary utility of unsupervised learning is to guide search away from unpromising sequences in the fitness landscape based on a (semantically meaningful) sense of their unnaturalness.
Pretty cool! But it also suggests that unsupervised learning alone isn& #39;t enough to discover enhanced, >WT variants.
So could this be what the low-N training mutants are for?
Spoiler ... looks like it!
Spoiler ... looks like it!
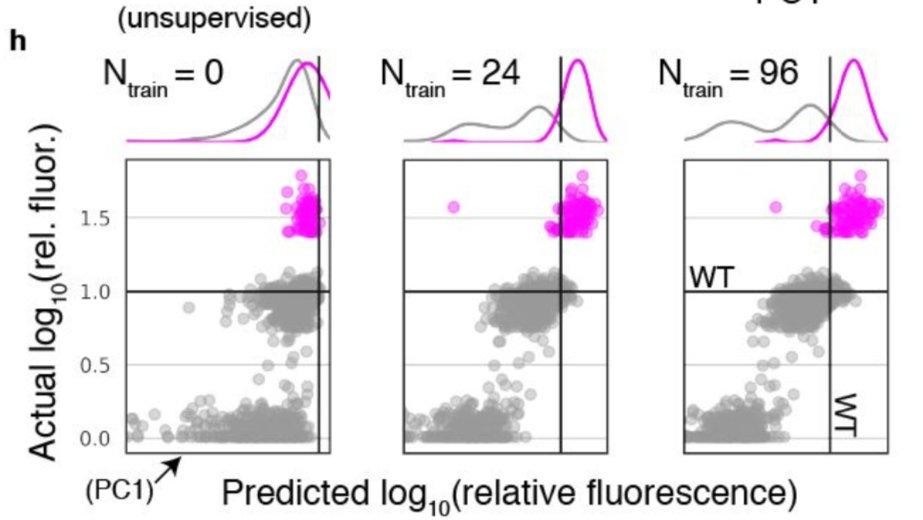
To see this, first note that PC1 is just a weighted combination of elements in the protein representation vector, where the weights are learned in an unsup manner. With labeled data we can learn new weights, via (supervised) regression. This gives a new 1d summary of the protein
If we plot protein activity vs this 1 dimensional summary across increasing amounts of low-N training data, we see that low-N supervision is necessary to be able to differentiate enhanced, >WT variants from those with ~WT activity.
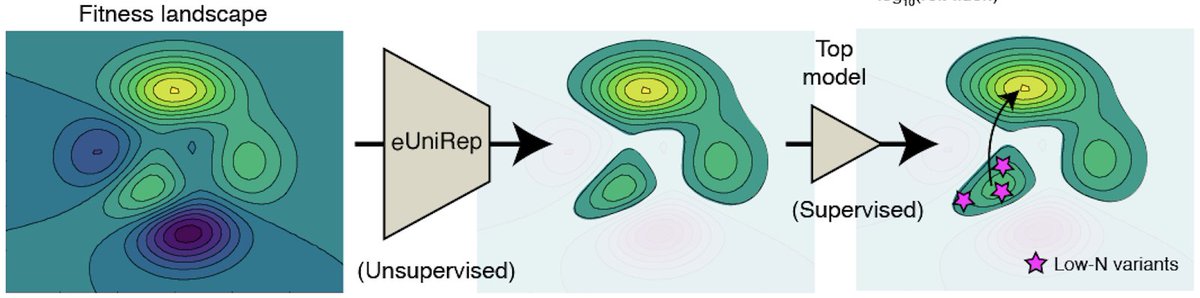
Putting it all together, we propose a simple two part model to explain why low-N ML guided protein design works.
1) Unsupervised learning simplifies search by eliminating the large swaths of the non-functional fitness landscape on the basis of unnaturalness.
2) “On top” of this information, supervised learning with a small number of low-N mutants then distills the information needed to discover better-than-natural variants.
Of course, there is a lot more work to be done to determine the generalizability of the above across different modeling & training paradigms and different classes of proteins ...
but, it may a provide some explanation for a puzzling result that the field has been seeing recently; that purely unsup ML-guided protein eng approaches have generally been unable to find seqs with >WT activity. It could be that some amount of supervision is needed!
Also, protein eng is as much figuring out what seqs NOT to spend time on as much as it is figuring out what seqs are worth exploring further. An unsupervised "filter" charged with a semantically meaningful notion of unnaturalness could be useful for the former.
Finally, we did a lot more to work to eliminate other hypotheses that could explain why low-N protein engineering works. The V2 update of our low-N paper contains new text and 5 new supp figures that explores these more deeply.
Check it out and let us know what you think! https://bit.ly/3jAV4ih ">https://bit.ly/3jAV4ih&q...

 Read on Twitter
Read on Twitter