Where is RNA in the bacterial cytoplasm? Well, it depends on the sequence and on the species—once more, there are not general rules  https://abs.twimg.com/emoji/v2/... draggable="false" alt="😱" title="Vor Angst schreiendes Gesicht" aria-label="Emoji: Vor Angst schreiendes Gesicht"> https://onlinelibrary.wiley.com/doi/abs/10.1002/wrna.1615">https://onlinelibrary.wiley.com/doi/abs/1...
https://abs.twimg.com/emoji/v2/... draggable="false" alt="😱" title="Vor Angst schreiendes Gesicht" aria-label="Emoji: Vor Angst schreiendes Gesicht"> https://onlinelibrary.wiley.com/doi/abs/10.1002/wrna.1615">https://onlinelibrary.wiley.com/doi/abs/1...
BTW one more nail in the coffin of the textbook assertion—mostly based on data on some genes of E. coli—that transcription/translation coupling & Rho-dep termination is a must for avoiding transcript chaos in the non-compartimentalized prok cytoplasm. Other bugs do differently  https://abs.twimg.com/emoji/v2/... draggable="false" alt="💪🏻" title="Angespannter Bizeps (heller Hautton)" aria-label="Emoji: Angespannter Bizeps (heller Hautton)">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="💪🏻" title="Angespannter Bizeps (heller Hautton)" aria-label="Emoji: Angespannter Bizeps (heller Hautton)">
I find this concept a most inspiring part of Orna Amster-Choder’s review: Chemical properties of given RNA sequences may drive formation of well-separated subcellular domains  https://abs.twimg.com/emoji/v2/... draggable="false" alt="➡️" title="Pfeil nach rechts" aria-label="Emoji: Pfeil nach rechts"> physical clustering of RNA is a cause, not the effect of a distinct 3D cytoplasmic architecture
https://abs.twimg.com/emoji/v2/... draggable="false" alt="➡️" title="Pfeil nach rechts" aria-label="Emoji: Pfeil nach rechts"> physical clustering of RNA is a cause, not the effect of a distinct 3D cytoplasmic architecture  https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Rückhand Zeigefinger nach unten" aria-label="Emoji: Rückhand Zeigefinger nach unten">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Rückhand Zeigefinger nach unten" aria-label="Emoji: Rückhand Zeigefinger nach unten">

 Read on Twitter
Read on Twitter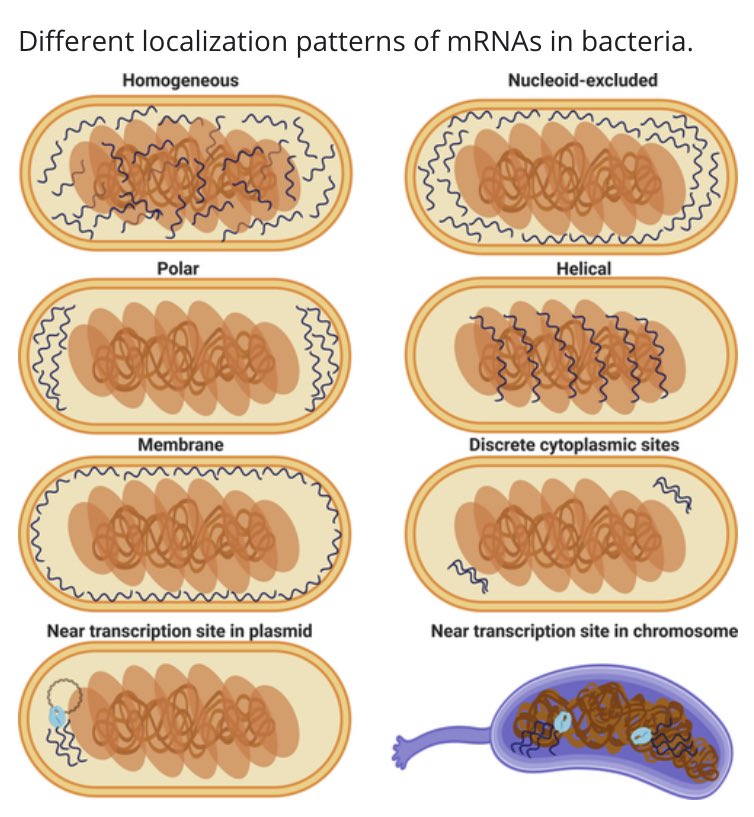 https://onlinelibrary.wiley.com/doi/abs/1..." title="Where is RNA in the bacterial cytoplasm? Well, it depends on the sequence and on the species—once more, there are not general rules https://abs.twimg.com/emoji/v2/... draggable="false" alt="😱" title="Vor Angst schreiendes Gesicht" aria-label="Emoji: Vor Angst schreiendes Gesicht"> https://onlinelibrary.wiley.com/doi/abs/1..." class="img-responsive" style="max-width:100%;"/>
https://onlinelibrary.wiley.com/doi/abs/1..." title="Where is RNA in the bacterial cytoplasm? Well, it depends on the sequence and on the species—once more, there are not general rules https://abs.twimg.com/emoji/v2/... draggable="false" alt="😱" title="Vor Angst schreiendes Gesicht" aria-label="Emoji: Vor Angst schreiendes Gesicht"> https://onlinelibrary.wiley.com/doi/abs/1..." class="img-responsive" style="max-width:100%;"/>
 physical clustering of RNA is a cause, not the effect of a distinct 3D cytoplasmic architecture https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Rückhand Zeigefinger nach unten" aria-label="Emoji: Rückhand Zeigefinger nach unten">" title="I find this concept a most inspiring part of Orna Amster-Choder’s review: Chemical properties of given RNA sequences may drive formation of well-separated subcellular domains https://abs.twimg.com/emoji/v2/... draggable="false" alt="➡️" title="Pfeil nach rechts" aria-label="Emoji: Pfeil nach rechts"> physical clustering of RNA is a cause, not the effect of a distinct 3D cytoplasmic architecture https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Rückhand Zeigefinger nach unten" aria-label="Emoji: Rückhand Zeigefinger nach unten">" class="img-responsive" style="max-width:100%;"/>
physical clustering of RNA is a cause, not the effect of a distinct 3D cytoplasmic architecture https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Rückhand Zeigefinger nach unten" aria-label="Emoji: Rückhand Zeigefinger nach unten">" title="I find this concept a most inspiring part of Orna Amster-Choder’s review: Chemical properties of given RNA sequences may drive formation of well-separated subcellular domains https://abs.twimg.com/emoji/v2/... draggable="false" alt="➡️" title="Pfeil nach rechts" aria-label="Emoji: Pfeil nach rechts"> physical clustering of RNA is a cause, not the effect of a distinct 3D cytoplasmic architecture https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Rückhand Zeigefinger nach unten" aria-label="Emoji: Rückhand Zeigefinger nach unten">" class="img-responsive" style="max-width:100%;"/>


