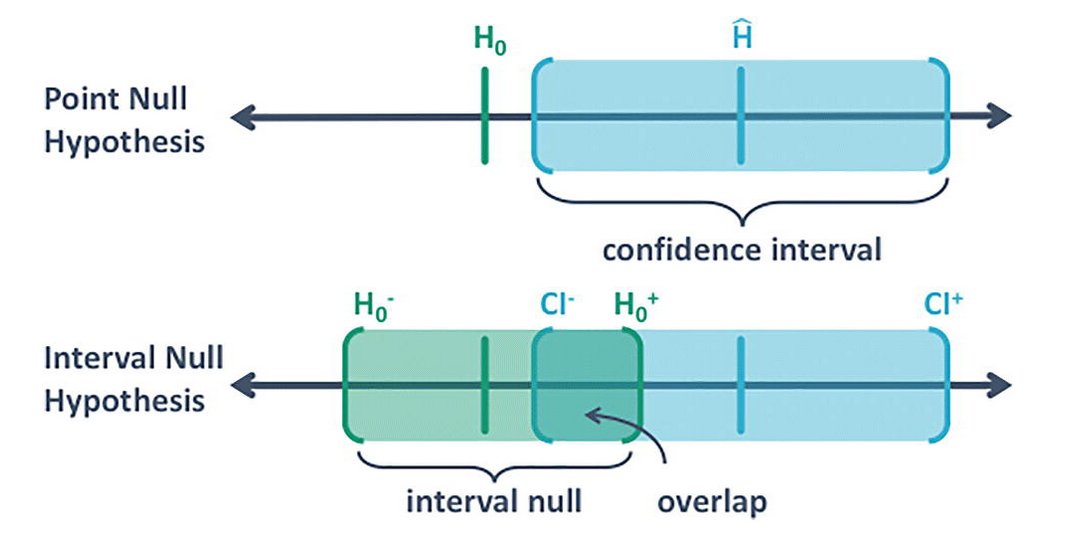
I just learned about second-generation p-values, by @StatEvidence.
They make conceptually more sense as they take into account that the null has an interval too. That there is a difference between null and practically null.
1/
h/t @f2harrell
https://www.tandfonline.com/doi/full/10.1080/00031305.2018.1537893">https://www.tandfonline.com/doi/full/...
They make conceptually more sense as they take into account that the null has an interval too. That there is a difference between null and practically null.
1/
h/t @f2harrell
https://www.tandfonline.com/doi/full/10.1080/00031305.2018.1537893">https://www.tandfonline.com/doi/full/...
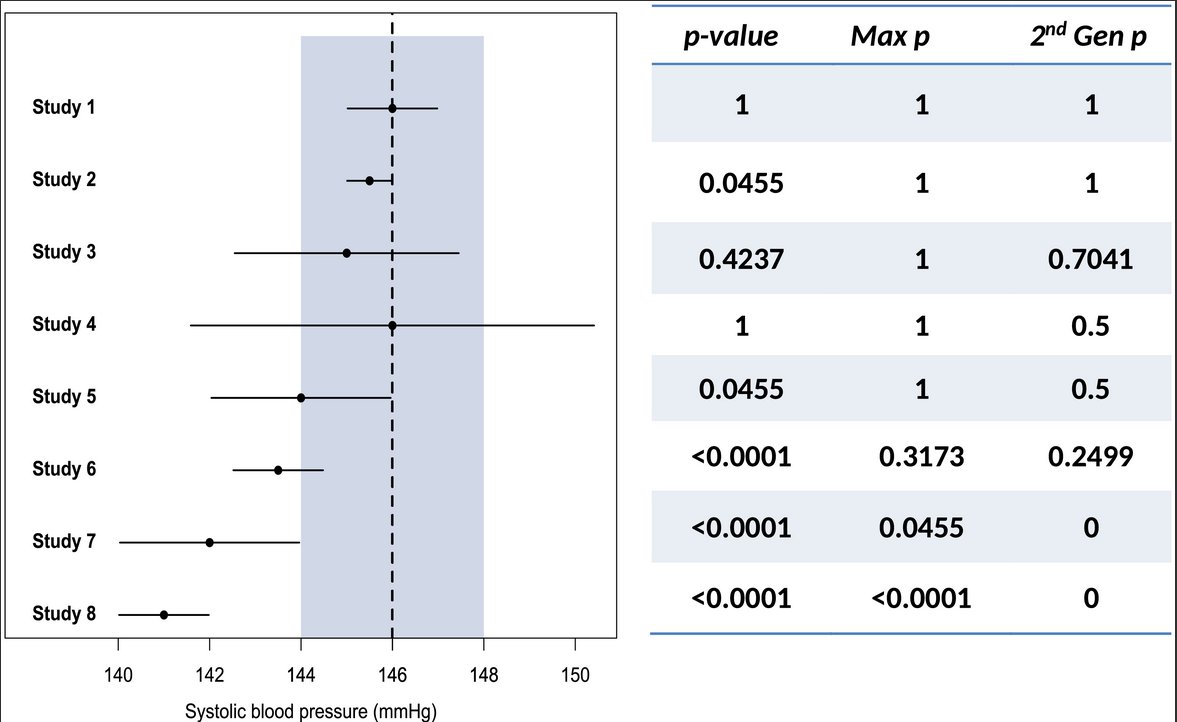
See how they are applied, with the Interval Null in grey.
https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0188299">https://journals.plos.org/plosone/a...
https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0188299">https://journals.plos.org/plosone/a...
Here& #39;s the method applied to gene expression data
This method puts a reality check on research, which is particularly relevant when using ultra-large datasets in which small effect sizes are flagged statistically significant, while rare variants with larger effect sizes still fail to make the cut.
Many researchers may not like that you have to specify the interval thresholds, think that is too subjective, but I think that is a conversation we desperately need. With datasets getting larger, we need to discuss: when are effect sizes *practically* null.
/end
/end

 Read on Twitter
Read on Twitter