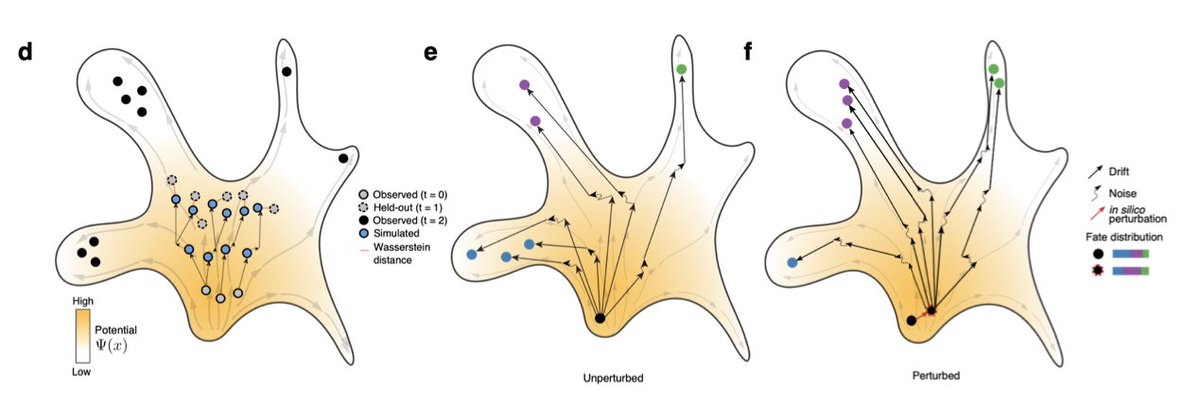
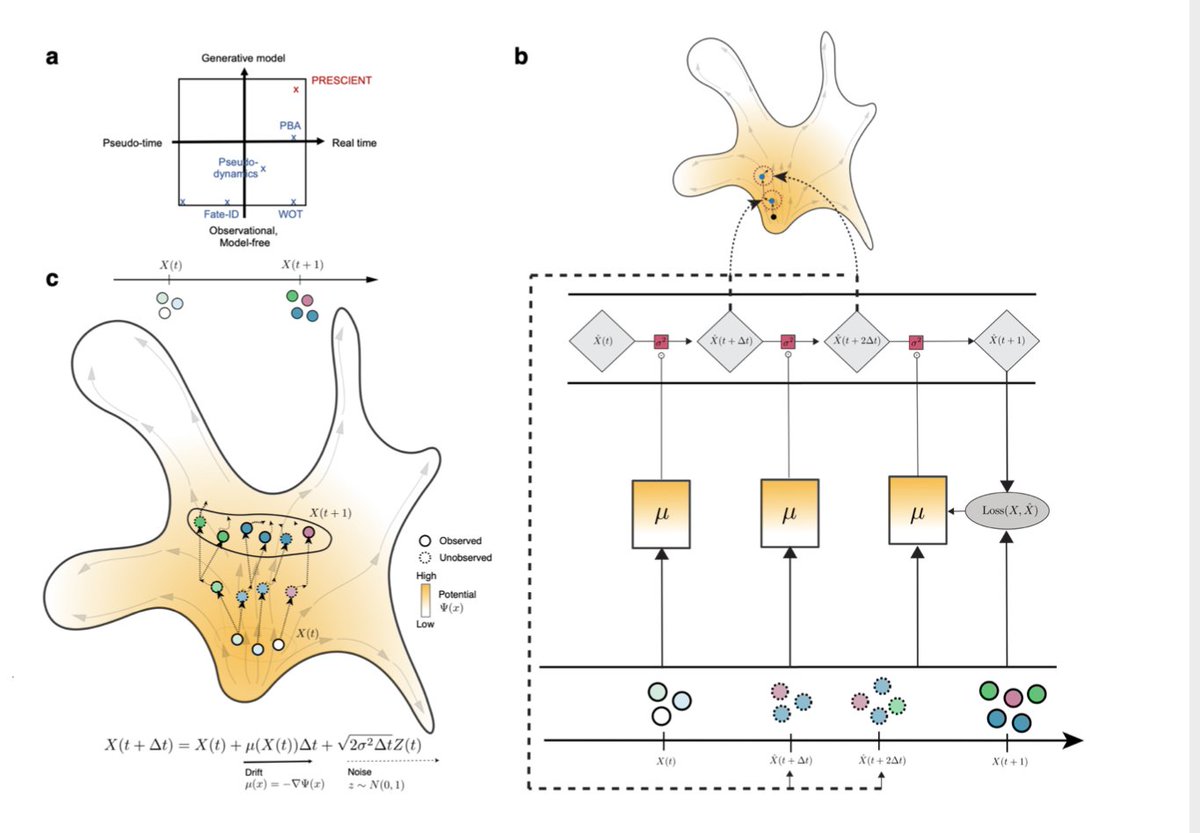
PRESCIENT is a generative modeling framework for longitudinal scRNA-seq datasets that was just released by the Gifford group at @MIT_CSAIL.
Applications include:
-predicting cell fate
-predicting consequences of in silico perturbations
h/t @msbr89
1/n
Applications include:
-predicting cell fate
-predicting consequences of in silico perturbations
h/t @msbr89
1/n
This is not another of the 70+ trajectory inference methods that try to infer a developmental ordering of the cell from a single experiment (see this @NatureBiotech article).
Instead, PRESCIENT is for when you have scRNA-seq from different time points. https://www.nature.com/articles/s41587-019-0071-9">https://www.nature.com/articles/...
Instead, PRESCIENT is for when you have scRNA-seq from different time points. https://www.nature.com/articles/s41587-019-0071-9">https://www.nature.com/articles/...
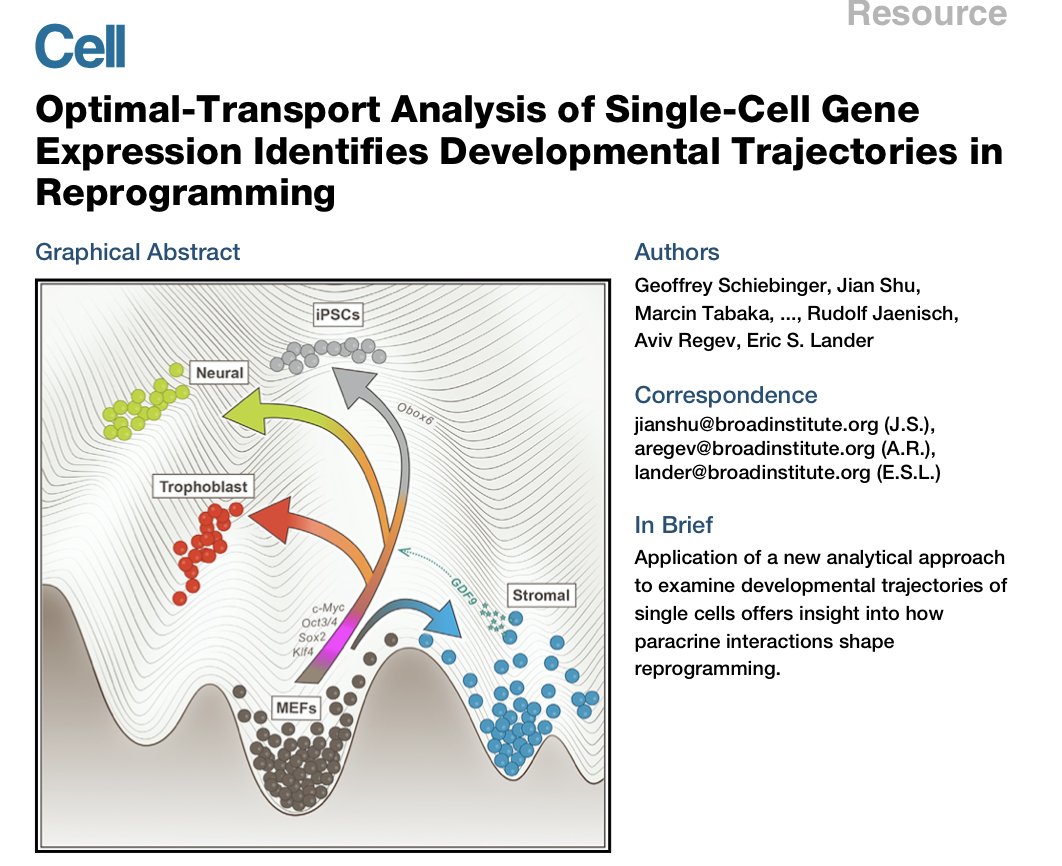
Previous work in this setting includes a beautiful paper from @geoffschieb, Regev, @eric_lander, and others, which considers cells as moving according to a potential function. Trajectories can then be identified using optimal transport. But it& #39;s not a generative model.
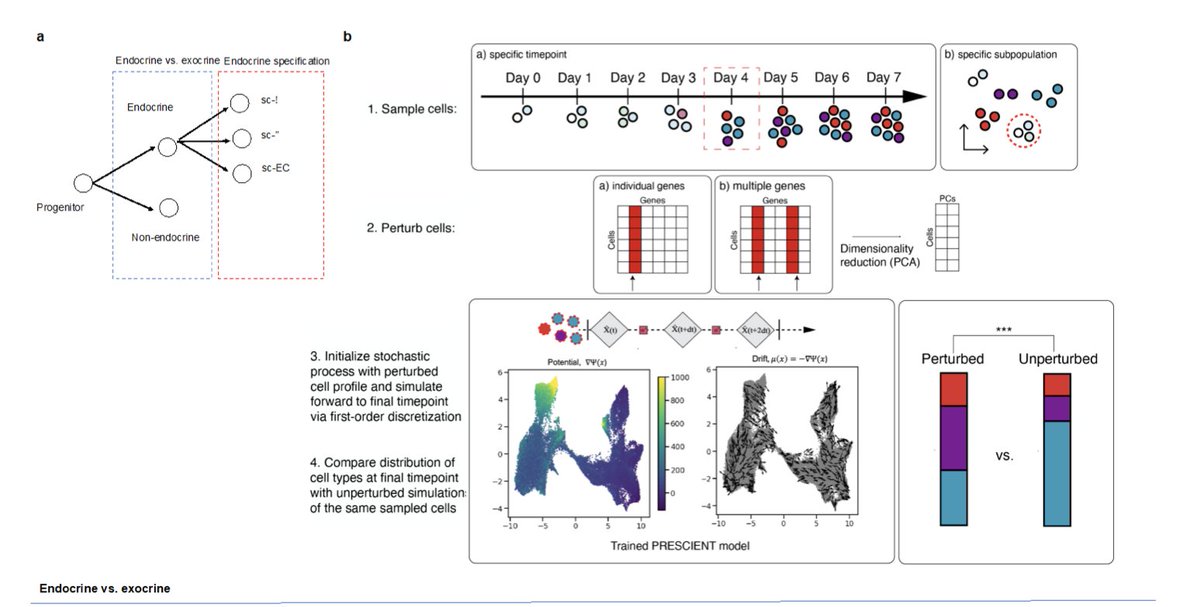
In contrast, PRESCIENT parameterizes the potential function itself using a neural network. Each cell is modeled as moving according to this potential, minimizing the error at each observed time step.
This potential function can then be used to sample new points/trajectories.
This potential function can then be used to sample new points/trajectories.
But the process needs to account for cell proliferation across time. The authors modified an approach from the Schiebinger paper above, which models a birth-death process, fit based on the expression of genes annotated as birth (KEGG_CELL_CYCLE) or death (KEGG_APOPTOSIS).
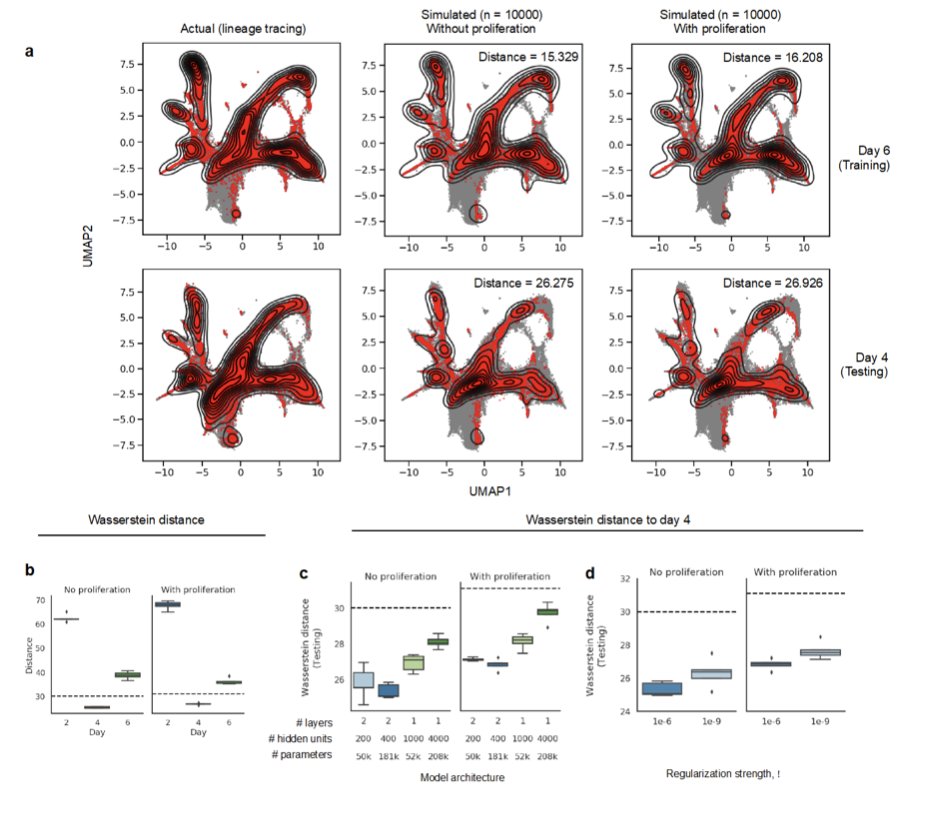
To validate, they used lineage tracing from @caleb_weinreb, @KleinLabHMS, et al, where clones are tagged and then sequenced at various time points, tracking their development (LARRY).
When trained on days 2 and 6, PRESCIENT predicts day 4 (but does not look too accurate to me).
When trained on days 2 and 6, PRESCIENT predicts day 4 (but does not look too accurate to me).
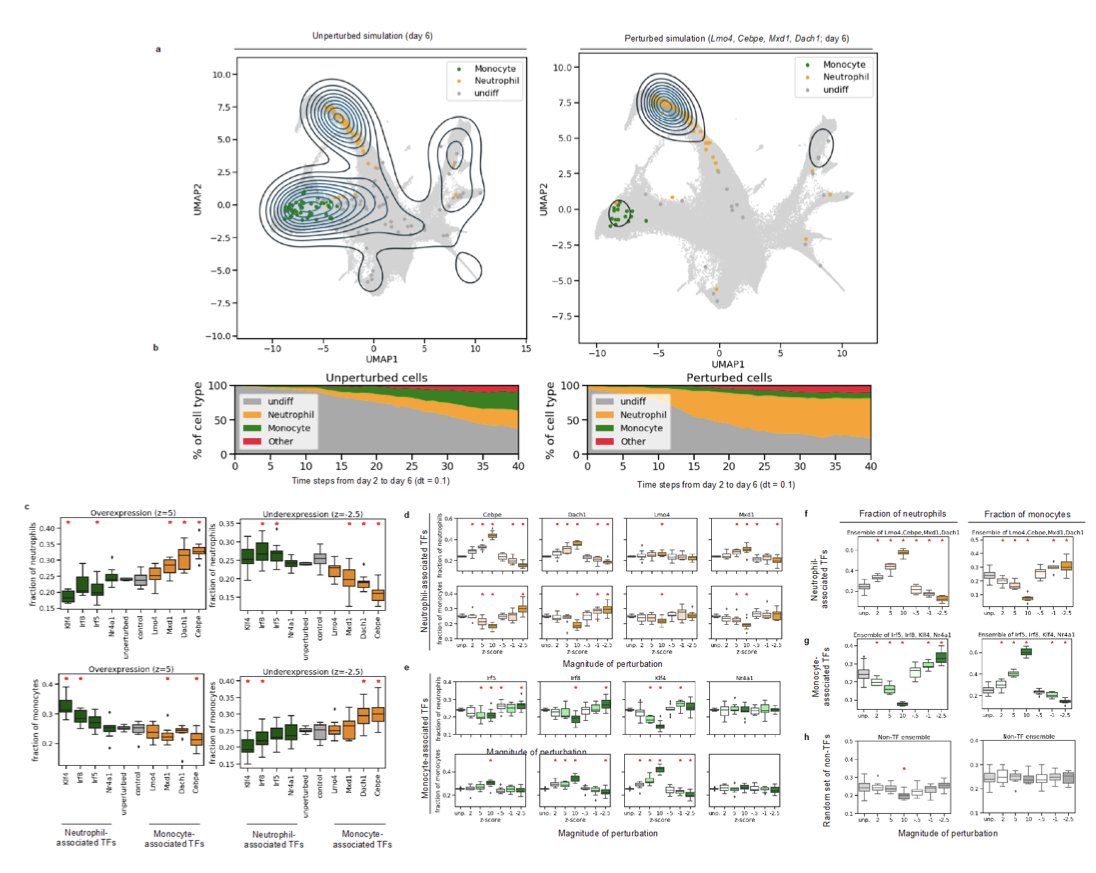
The in-silico perturbation experiments were most interesting.
They increased the expression of genes known to be involved in neutrophil differentiation, and the model accordingly produced more mature neutrophils. Same with monocytes.
They increased the expression of genes known to be involved in neutrophil differentiation, and the model accordingly produced more mature neutrophils. Same with monocytes.
Then to really demonstrate the power of these in silico experiments, they did a screen of 200+ TFs, to see which will alter cell fate in induction of human pancreatic islet cells. They identified 28 TFs, some of which had been previously shown to have the observed effect.
Overall, while much more work needs to be done, generative modeling of scRNA-seq may be very useful for in silico perturbation analyses--and presents some interesting computational problems.
Paper: https://www.biorxiv.org/content/10.1101/2020.08.26.269332v1
Code:">https://www.biorxiv.org/content/1... https://github.com/gifford-lab/prescient">https://github.com/gifford-l...
Paper: https://www.biorxiv.org/content/10.1101/2020.08.26.269332v1
Code:">https://www.biorxiv.org/content/1... https://github.com/gifford-lab/prescient">https://github.com/gifford-l...

 Read on Twitter
Read on Twitter