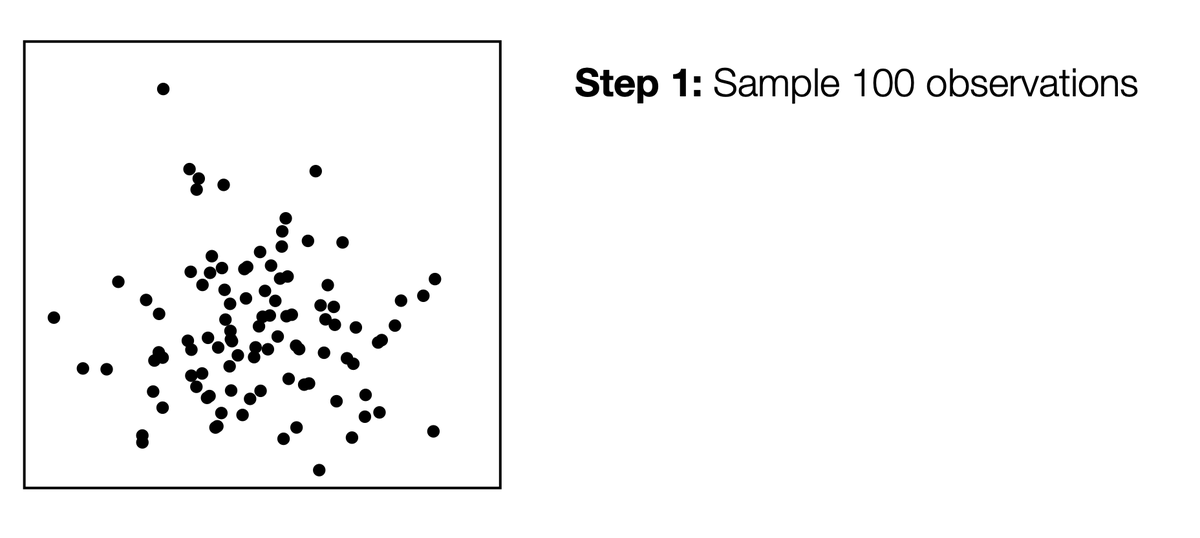
"Double-dipping" - generating a hypothesis based on your data, and then testing the hypothesis on that same data - is dangerous. To see this, let& #39;s take data with no signal at all ... 1/
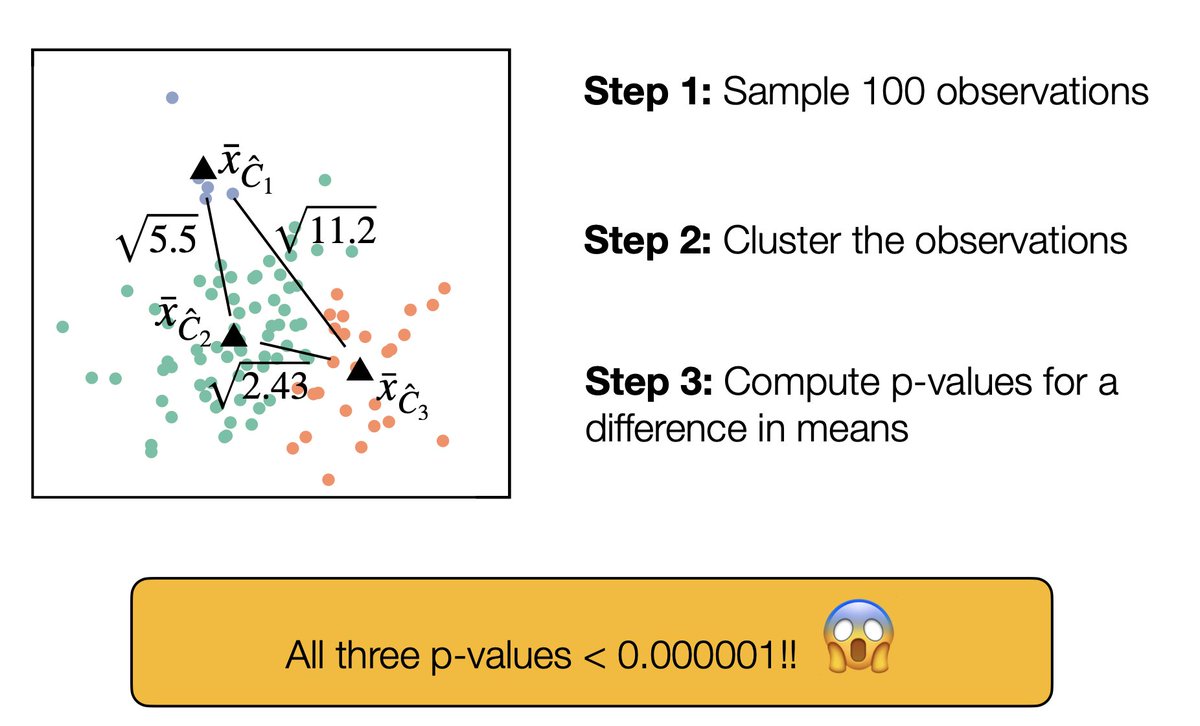
... Cluster that data set, then compute p-values for a difference in means between the clusters. The p-values are tiny, even though none of the clusters are real!!! 2/
Can we fix this by sample splitting? Let& #39;s divide our data into two parts, and this time only cluster Part 1. We& #39;ll label Part 2 using a classifier trained on Part 1, then compute p-values for a difference in means on Part 2. 3/
This might seem ok, because we used different data to cluster and test ... but the p-values are STILL tiny! Don& #39;t believe me? Try it for yourself at https://tinyurl.com/y2cw2rlg !">https://tinyurl.com/y2cw2rlg&... 4/
What& #39;s going on?! Well, sample splitting involves a sneaky form of double dipping. Labelling your test set counts as using it to generate a null hypothesis. Then, you use the test set to test that null hypothesis. 5/
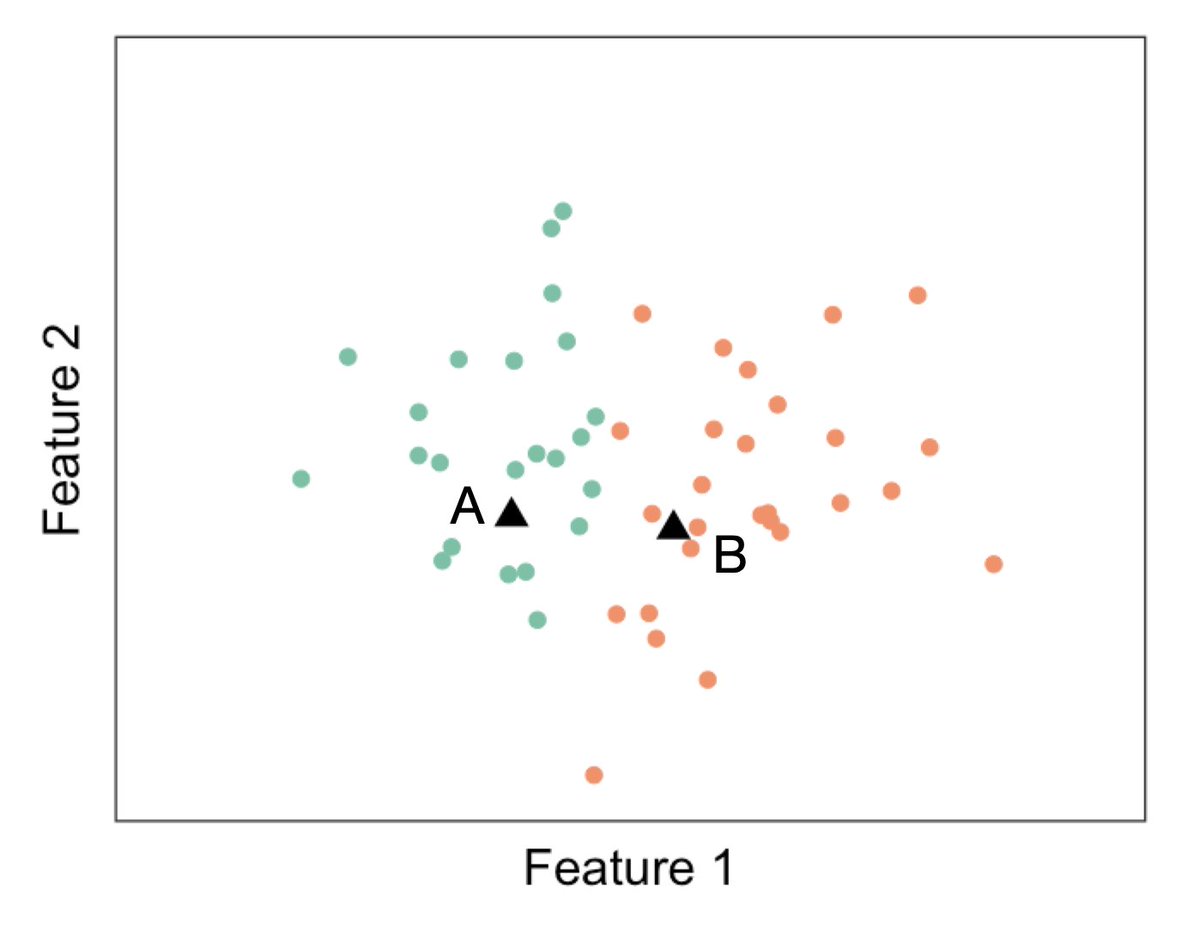
Here& #39;s another sneaky one! let& #39;s say we 1) Randomly generate Datapt A and Datapt B. 2) Call our observations green if they& #39;re closer to DataPt A, and orange if they& #39;re closer to DataPt B. 3) Test for a difference in means between green and orange. Is this double dipping??? 5/
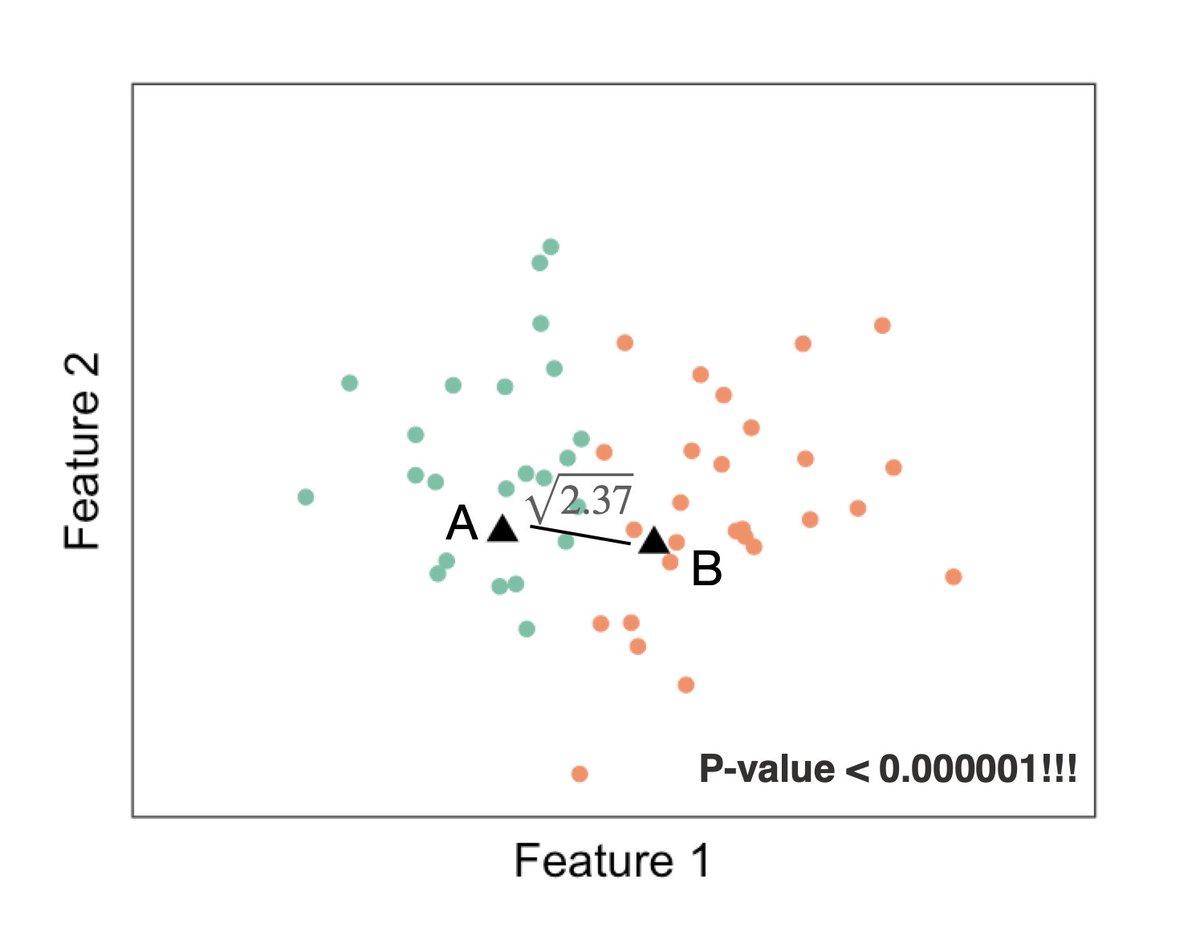
Yep, double dipping is back again ... by assigning our observations to different groups, we used our data to generate a null hypothesis, then we use the same data to test it. Once again the p-value is tiny. 6/
The good news is that we (me, @daniela_witten & Jacob Bien) have a fix for double-dipping for clustering!! The key idea is to adjust for double dipping by conditioning on the hypothesis generation procedure. 7/
We& #39;re working hard on getting the paper out! My talk on this topic is also available at https://drive.google.com/file/d/1iljgZzjfjbJNnmbCTbwp91vGiVhfvsyw/view?usp=sharing">https://drive.google.com/file/d/1i... :) 8/END

 Read on Twitter
Read on Twitter