If pleiotropy is simply when one mutation influences many traits, how come so many nuanced definitions of it exist? 1/17
One answer is that it is not easy to count traits. For example, consider cell circumference and area. Are those two traits or one? For a harder example, consider the mutation that makes tomatoes ripen uniformly and also makes them taste bad. Sounds like pleiotropy, right? 2/17
But is it fair to call this tomato mutation pleiotropic if its singular effect is to reduce function of a transcription factor that promotes chloroplast development, which in turn affects both coloration and sugar accumulation? (This is called ‘vertical pleiotropy’). 3/17
These questions about pleiotropy hint at deeper questions about the organization of biological systems: to what extent are the phenotypes of an organism interconnected and dependent on one another, and to what extent are traits independent at the molecular and higher levels? 4/17
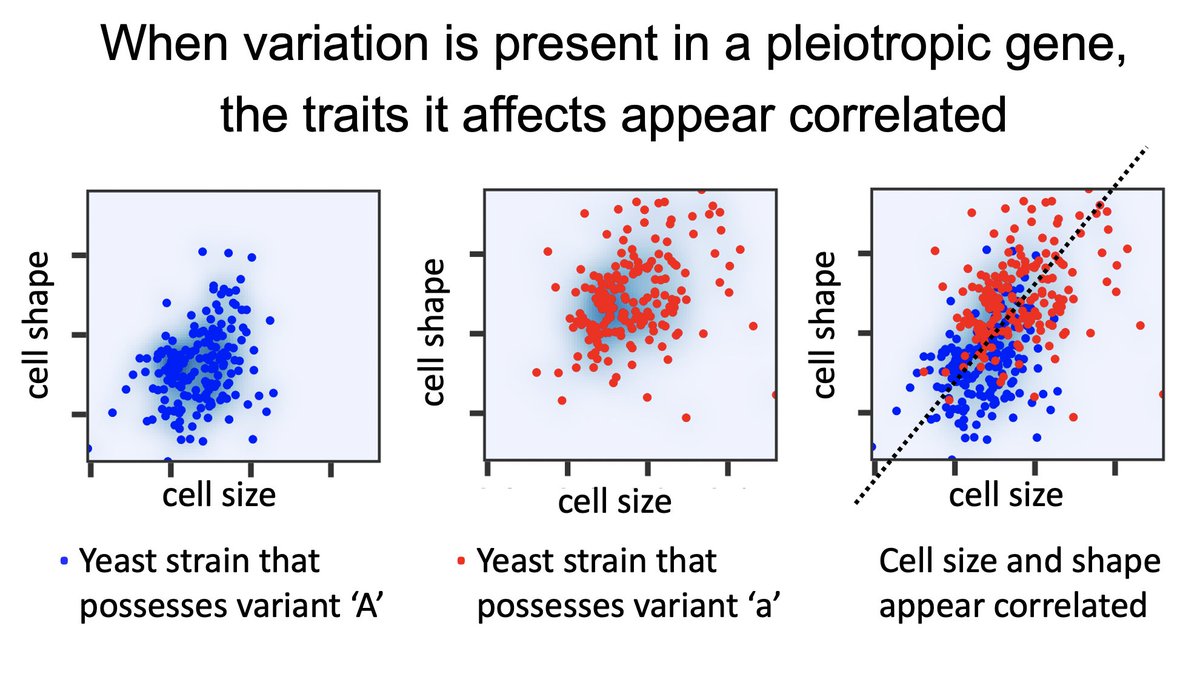
This question is hard to answer because pleiotropic genetic variants can cause independent traits to appear correlated! Below, cell size and shape are not so correlated in either subpopulation, but more correlated when there is variation present in the pleiotropic gene. 5/17
If we could quantify the relationships that exist between traits in the absence of genetic variation, it might improve predictions about the genotype-phenotype map, especially predictions about which traits are likely to be jointly affected by the same genetic change. 6/17
It would also help us understand the structure of biological systems, the degree to which their molecular and higher-level features are interconnected, how many of these features a typical mutation modifies at one time, and how this impacts the way these systems evolve. 7/17
In our new paper in @PLOSBiology, we move towards these goals. First, we quantify the inherent relationships between traits by studying them in clonal populations of yeast cells. w/ Mark Siegal @ShuangLi_NYU @chlazaris @ZivNaomi @annalisepaaby 8/17 https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3000836">https://journals.plos.org/plosbiolo...
Then we ask whether pleiotropic genetic variation present in nature tends to influence related or unrelated traits. 9/17
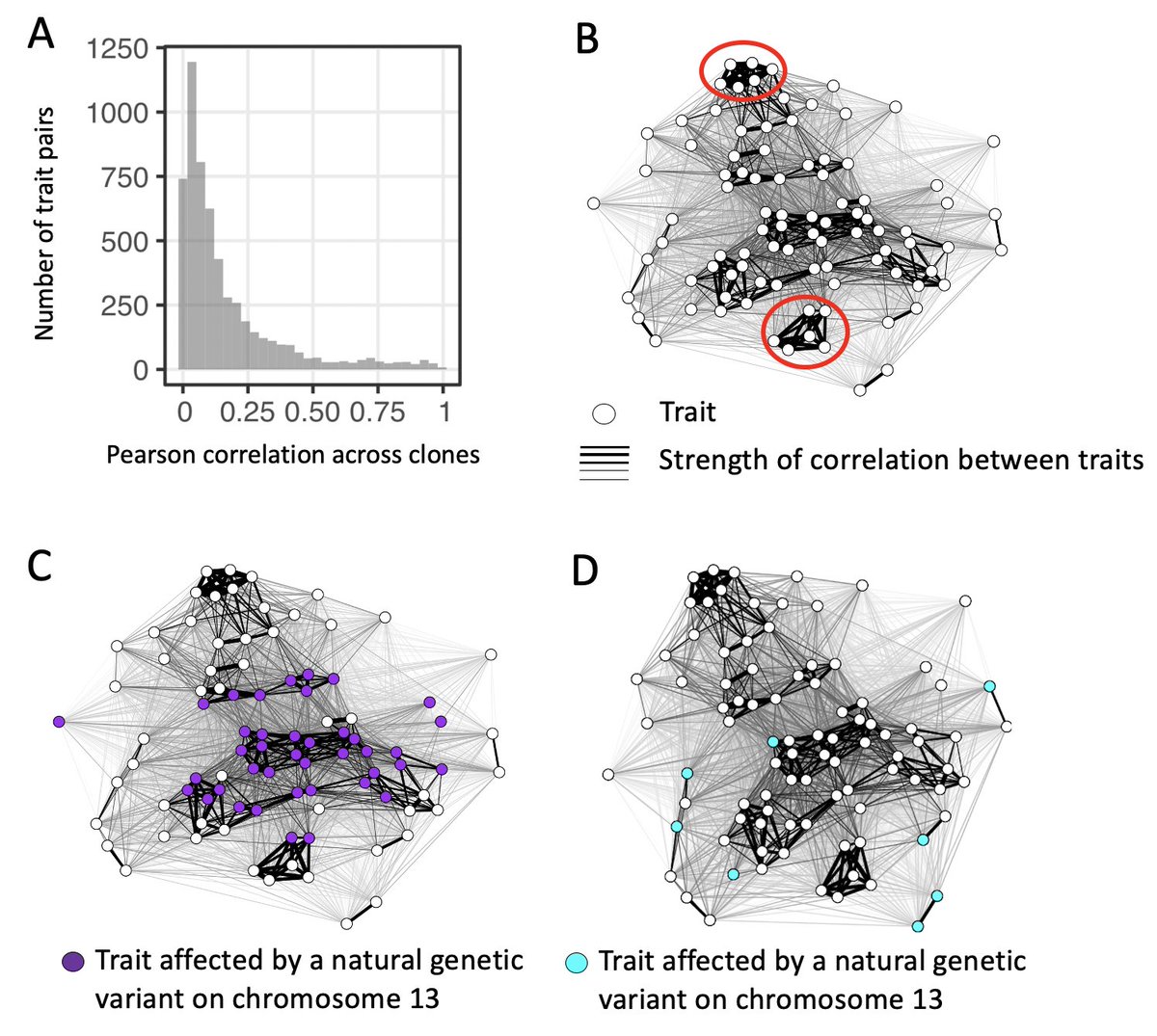
We find A) most of the ~150 single-cell morphological traits we study are not strongly correlated across clones, however B) groups of traits with strong correlations exist, C) pleiotropic alleles tend to influence correlated traits clusters, but D) sometimes they do not! 10/17
The observation that genetic variants present in nature sometimes influence many uncorrelated traits is one of our ‘big’ results. You might expect natural selection to purge such variants as they have greater potential to affect at least one trait in a negative way. 11/17
It’s possible that selection often purges this type of pleiotropic variant. A future study could determine whether this is the case by comparing our findings to those obtained when studying spontaneous mutations that have not been pruned by selection 12/17
Another finding of our paper – one that threw us for a loop and took several years and many headaches to sort out – is that the correlations between some traits change across the cell cycle, with genetic background, after drug treatment, and likely across other contexts. 13/17
This means that a gene might be pleiotropic (affect many traits) in some contexts, but not in other contexts where those traits are less correlated. This in turn means that the phenotypic effects of a genetic change may be hard to predict. 14/17
On a more optimistic note, this finding provides an explanation as to why selection may not purge pleiotropic variation. Such variants might not have had high pleiotropy in their original context. 15/17
Indeed, in another recent project with @PetrovADmitri and @GrantKinsler, we show that adaptive mutations have low pleiotropy in the environment where they evolved, but higher pleiotropy in other contexts. 16/17 https://www.biorxiv.org/content/10.1101/2020.06.25.172197v2">https://www.biorxiv.org/content/1...
To conclude, pleiotropy is – to me at least – a challenging concept, and it’s no surprise that many definitions exist. I hope that our new #PLOSBiology paper is helpful in explaining these challenges and takes a step forward in surmounting them. 17/17 https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3000836">https://journals.plos.org/plosbiolo...

 Read on Twitter
Read on Twitter