Remember March, the endless month? That was the month that the #covid19 #pandemic became real, at least where I work @UMassMedical @broadinstitute. Suddenly we were working from home, and a bit lost. So ... this new paper happened. Here& #39;s the story. 1/n https://www.pnas.org/content/early/2020/08/20/2010146117">https://www.pnas.org/content/e...
On March 19, Harris Lewin @EBPgenome sent me an email. He and @joana_damas had an idea. Could we use #genomics to predict which species were likely to be susceptible to #SARSCoV2 infection, the virus that causes #COVID19? 2/n
On April 18, we posted a preprint of our paper on @biorxivpreprint
This is really fast for science. We were analyzing and conferencing and slacking all hours of day, some of us zooming in our fabulous pajamas (looking at you @corrie_painter)
Why the urgency? 3/n
This is really fast for science. We were analyzing and conferencing and slacking all hours of day, some of us zooming in our fabulous pajamas (looking at you @corrie_painter)
Why the urgency? 3/n
Lots of scientists study humans. A few scientists study a few other species, but for most species we have no information on. And animals were getting sick. 4/n https://www.nytimes.com/2020/04/06/nyregion/bronx-zoo-tiger-coronavirus.html">https://www.nytimes.com/2020/04/0...
What about the animals in our zoos, or on ecotours, who might be exposed to infected people? What about the #Endangered species who may not survive another blow? Could another species unknowingly act as a #virus reservoir, and pose a risk to us in the future? 5/n
GENOMICS AND PROTEIN MODELLING TO THE RESCUE!!! (possibly the only time I will write that). @joana_damas @keoughkath @corrie_painter @hillermich @apfenning @frozenzoo @MNweeia @EmmaTeeling1 @KltLab, others. meeting by @zoom_us / talking via @SlackHQ / writing @googledocs 6/n
the logic:
In order to infect our cells, the virus #SARSCoV2 makes a protein called Spike. Spike binds to human protein called ACE2, and sneaks into our cells. Explained beautifully by @13pt @carlzimmer 7/n https://www.nytimes.com/interactive/2020/03/11/science/how-coronavirus-hijacks-your-cells.html">https://www.nytimes.com/interacti...
In order to infect our cells, the virus #SARSCoV2 makes a protein called Spike. Spike binds to human protein called ACE2, and sneaks into our cells. Explained beautifully by @13pt @carlzimmer 7/n https://www.nytimes.com/interactive/2020/03/11/science/how-coronavirus-hijacks-your-cells.html">https://www.nytimes.com/interacti...
the ACE2 protein is made by a gene, also called ACE2. 8/n
ACE2 is an important gene with many functions. Like most such genes, it is ancient. All vertebrates have a version of ACE2, with subtle differences. There is a human ACE2, a dog ACE2, a cat ACE2, a narwhal ACE2, a chicken ACE2, a pufferfish ACE2, a frog ACE2 ... 9/n
#SARSCoV2 Spike matches really, really, really, really well to *human ACE2*. Thus the #pandemic 10/n https://www.worldometers.info/coronavirus/ ">https://www.worldometers.info/coronavir...
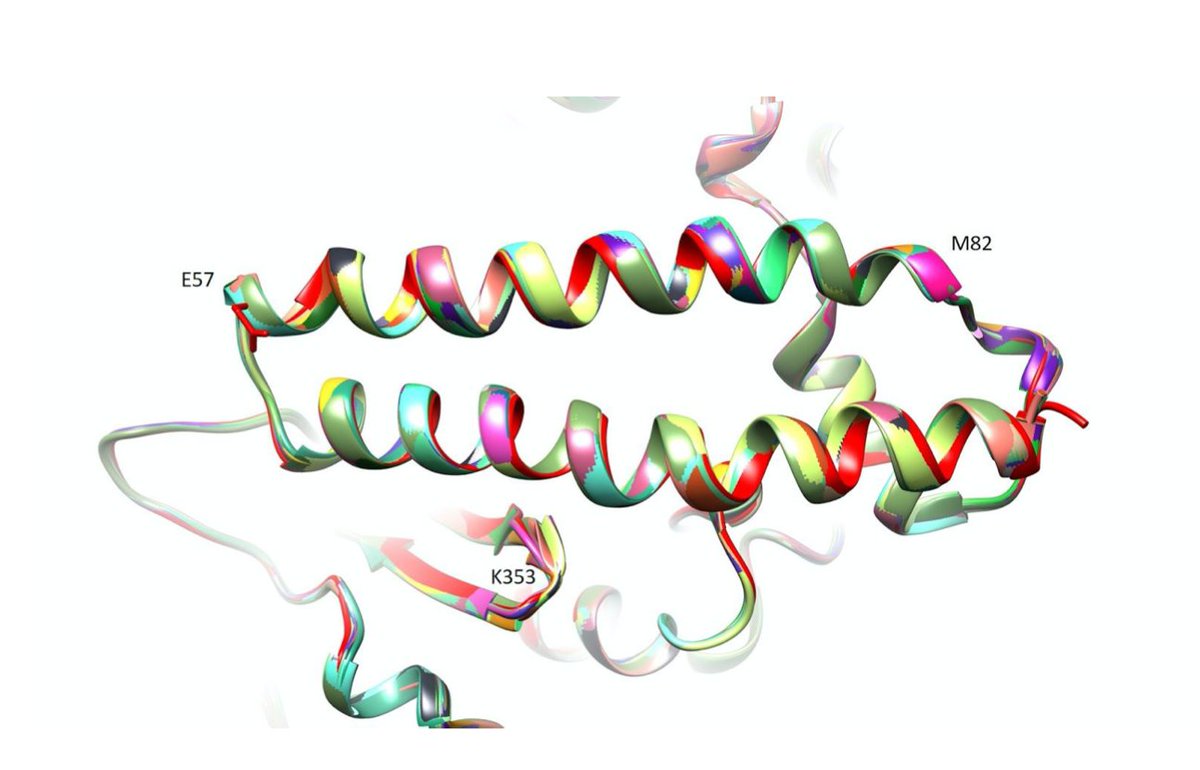
We assessed susceptibility to #SARSCoV2 by comparing human ACE2 (sequence and structure) to the ACE2 in 409 difference species. The more similar they were to human ACE2, the higher the risk. We focused on the part of ACE2 that binds to Spike (image from PNAS paper) 11/n
Disclaimer: this is an imperfect prediction. Lots of reasons it might not be accurate, as described in our paper.
To validate, we need more infectivity data, but that will take time (and never for most species).
And the pandemic is happening now. 12/n
To validate, we need more infectivity data, but that will take time (and never for most species).
And the pandemic is happening now. 12/n
This is where I give a HUGE shout out to #OpenData #OpenScience @NCBI. From home, in the middle of a pandemic, entirely virtually, we analyzed data from 410 species (inc humans). 252 mammals, 72 birds, 65 fishes, 17 reptiles, and 4 amphibians. Data shared by other scientists 13/n
and the @Genome10K affiliated genomics initiatives, including Zoonomia project (needs twitter handle! led by @KltLab and @karlssonlab) @bat1kgenomes @genomeark 14/n
We scored the 410 species into 5 risk categories, from very low to very high. The species scored as highest risk were all Old-World primates, including humans. Only mammals fell into the medium to very high categories. There were some surprises within mammals, though. 15/n
Full details in our #openaccess paper! 16/n https://www.pnas.org/content/early/2020/08/20/2010146117">https://www.pnas.org/content/e...
What’s next? with support from @NSF RAPID grant we’re now working to make predictions stronger- including more genes, and new infectivity data as it emerges. Stay tuned! 17/n

 Read on Twitter
Read on Twitter