In this week& #39;s "ML pitfalls w/ Jacob," we& #39;re going to talk about data set creation! Problem data sets occur in every field, but I frequently see them in genomics because people build their own data sets from new experimental data. 1/
The key idea: if you aren& #39;t careful about how you construct your data set, you may inject artificial (and hidden!) structure/bias that renders it useless even though models may appear to perform well. 2/
Let& #39;s consider a simple example. If you build a data set for car classification but it, accidentally, only contains blue SUVs and red pickups, a model can perform well by focusing on color and not car shape. 3/
Riberio et al. considered another example where they classify wolves (w/ snow background) vs huskies (w/ no snow). Using LIME on the model revealed that it focused on the background and ignored the dog. https://arxiv.org/abs/1602.04938 ">https://arxiv.org/abs/1602.... 4/
Data set issues can be difficult to track down. Feature attribution methods don& #39;t always work. If you apply those to the car example you& #39;d still see car pixels as important because those are the ones that have color! 5/
In the setting of genomics, a good example is the TargetFinder data set, which contained gene-enhancer links (and non-links) validated using 3D genome structure experments. https://www.nature.com/articles/ng.3539">https://www.nature.com/articles/... 6/
The authors faced two challenges when building this data: 1) in theory, the labels covered every pair of regions in the genome. That& #39;s too big to handle. 2) Most interactions are between regions close together on the genome. See this Hi-C matrix from wikipedia. 7/
To overcome these challenges they extracted a subset of contacts and controlled for distance, i.e. an equal # of contact/non-contact pairs at each distance so the model didn& #39;t simply learn "short range = contact." 8/
Sounds good, right? Unfortunately, most gene-enhancer pairs are not in contact and so controlling for distance led to negative examples having many repeats of the same genes but positive examples never having repeats. 9/
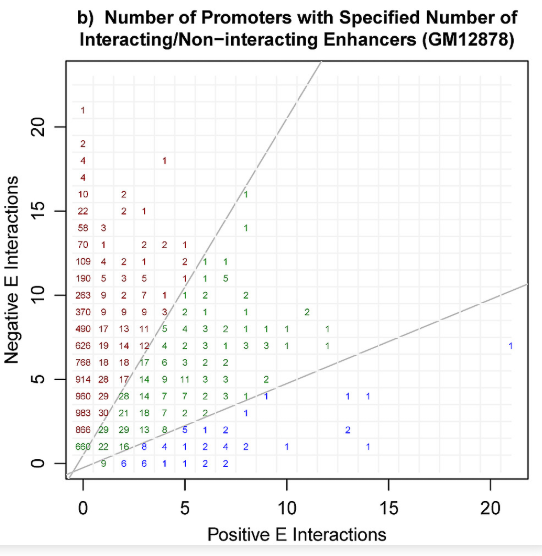
You can see here that 253 genes (promoters) were involved with 10 negative interactions and no positive interactions, and overall, most genes had far more negative interactions than positive ones. https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1006625">https://journals.plos.org/ploscompb... 10/
What& #39;s the problem? Well, if you just memorized the genes with many negative interactions and predicted negative for any interaction they were in, you& #39;d probably get great performance (all the reds in the previous figure). 11/
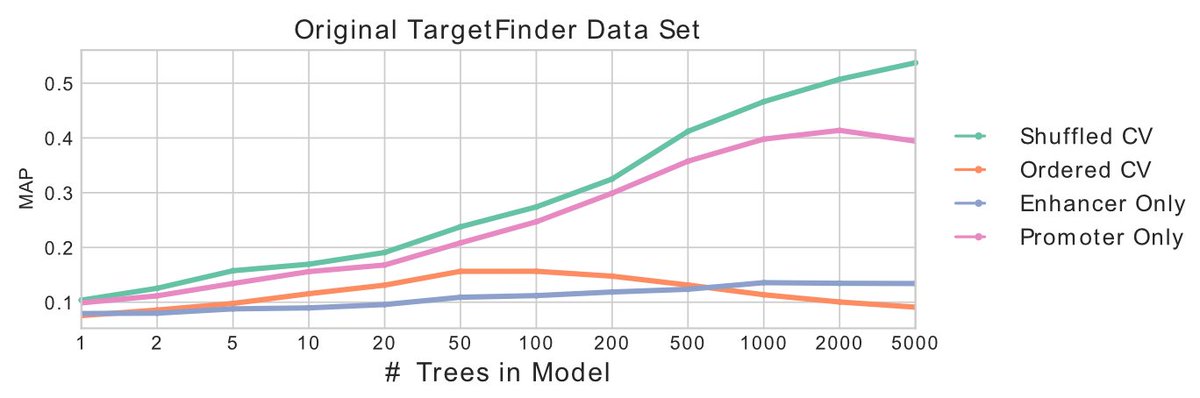
How do we know that& #39;s the case? Well, first clue: model performance *should* level off with complexity but we see that an XGBoost model *consistently* performs better with more trees, a big clue that memorization is happening. 12/
Second clue: when we train on some chromosomes and evaluate on other chromosomes this performance plummets. The model still memorized the negative genes, but those never appear in the test set so the predictions are poor. 13/
Third clue: When we use features from ONLY the gene and not the enhancer we get almost as good performance as using both. Doesn& #39;t seem realistic that you can predict gene-enhancer interactions without considering the enhancer! 14/
The problem was that their data set construction procedure made the identity of the genes (and not its features) predictve. Replacing the feature values with random numbers gives the exact same result. 15/
tl;dr when constructing your data sets make sure that you haven& #39;t accidentally introduced hidden structure into it. The easiest way to do this is make sure you CAN& #39;T predict things you shouldn& #39;t be able to. 16/
btw this thread is not meant to be a swipe at the authors of TargetFinder. When this issue came out last year they immediately agreed it was a problem and wrote about it. They did exactly what you& #39;re supposed to do and I& #39;m happy I get to collab w/ them. https://www.nature.com/articles/s41588-019-0473-0?proof=trueMay">https://www.nature.com/articles/...

 Read on Twitter
Read on Twitter