The more papers I read for a review article I& #39;m writing about ML pitfalls in genomics, the more my faith is shaken in the results from papers that apply machine learning to methylation arrays. A salty thread. 1/
A canonical mistake you can make when performing machine learning involves performing data preprocessing outside of cross-validation. This involves applying transformations or feature selections before splitting into a train/test split. 2/
In principle, being able to perform feature selection on methylation arrays is invaluable because they typically have 100ks of probes but only hundreds of examples. The newest EPIC arrays from Illumina have over 850k probes! 3/
In practice, around half of the papers I& #39;m reading involve calculating the predictive power of each probe individually across the entire data set, selecting the top X probes, and then performing cross-validation to evaluate performance. 4/
Frequently in the context of methylation arrays this process happens when people first identify "differentially methylated regions" (DMRs). These are probes that exhibit different signal across conditions, i.e. the labels they& #39;re about to predict. 5/
What& #39;s wrong with this? Well, you& #39;re leaking information from your test set into your training set because you& #39;re selecting probes that, by construction, have large differences / perform well on both your training and test set. 6/
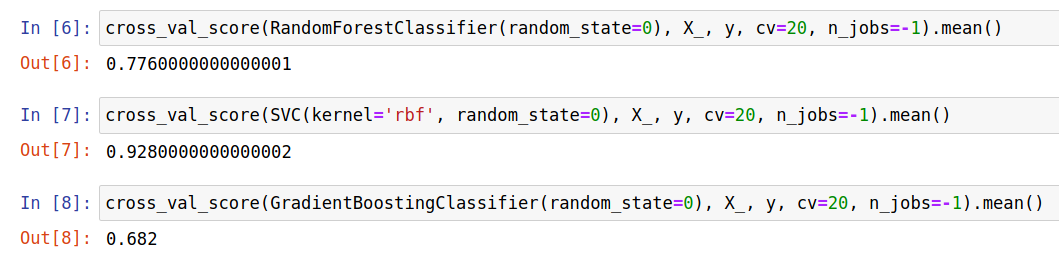
I& #39;ve used this example in the past: consider ENTIRELY RANDOM data. What happens if you select the top features and then do cross-validation? You get better than random performance because the selected features coincidentally line up with the labels. 7/
This isn& #39;t a problem that requires regularization. Everything is able to find signal despite the data being random. 8/
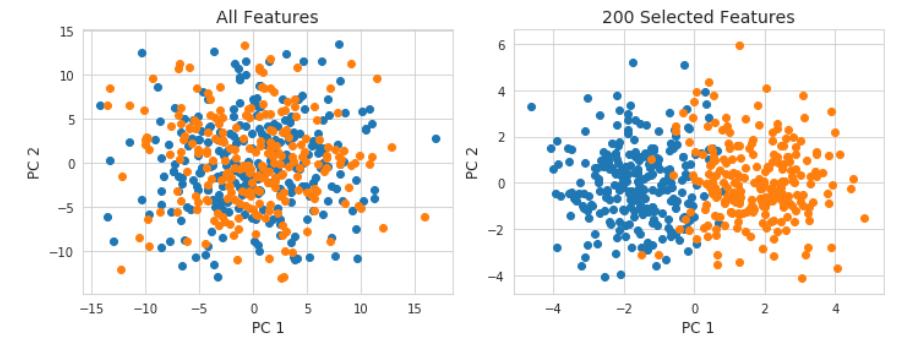
This also isn& #39;t a problem with supervised machine learning models. If you take your data, select down a smaller number of features (here going from 10k features to 200) even PCA will return distinct clusters. 9/
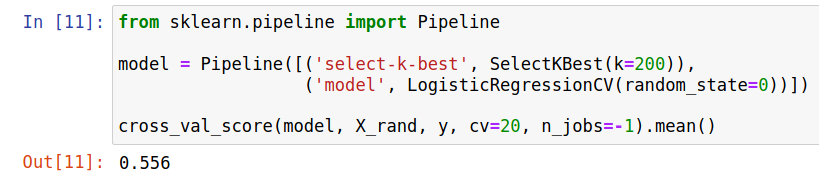
How do I know this has to do with data preprocessing being outside the train/test split and not me actually secretly generating a data set with secret structure in it? Let& #39;s put the feature selection IN the CV. Performance plummets. 10/
The inspiration for this thread was a methylation data analysis library that I don& #39;t think is necessary to name that was codifying both imputation of values and feature selection before the train/test split. 11/
Seeing this mistake in scientific papers is bad enough but seeing it be subtly integrated into workflows means even more people will inadvertently make this mistake. If you are working with methylation arrays, please ensure you do probe selection only on the training set! 12/12

 Read on Twitter
Read on Twitter