Just out: a piece of collaborative science I& #39;m rather proud of. Open data from @noahdsimons&co., a blog post from me, and epic analysis from Eerik, Uli and @adamjamesreid came together for this @PLoSPathogens tale of an unexpected genome.
( https://abs.twimg.com/emoji/v2/... draggable="false" alt="🧵" title="Thread" aria-label="Emoji: Thread">below..) https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1008717">https://journals.plos.org/plospatho...
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🧵" title="Thread" aria-label="Emoji: Thread">below..) https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1008717">https://journals.plos.org/plospatho...
(
For me the story (sorry if you& #39;ve heard it before) starts when I was looking into a malaria parasite gene I thought might be involved in gamete production, but which was annotated merely as "Conserved gene, unknown function".
I wondered if there was a related gene in another species that was better understood, so I decided to search for similar sequences in the NCBI database using BLAST.
As expected, the closest matches were in other species of malaria parasite. But nestled between the parasite sequences was a gene in a species I& #39;d never heard of: Piliocolobus tephrosceles.
I guessed it must be some obscure parasite, so was surprised when I put it into Wikipedia:
I guessed it must be some obscure parasite, so was surprised when I put it into Wikipedia:
Why would a monkey gene be so related to a gene from a malaria parasite? When I saw a similar result for a totally different malaria parasite gene I began to wonder if something more than evolution was afoot.
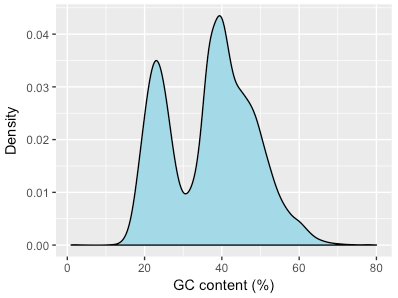
I downloaded the whole monkey genome assembly, and plotted the distribution of Gs and Cs vs As and Ts in the sequences that made it up.
There were two clear peaks. One was at 40% GC, similar to human genomes. But the other was at 20%, typical of malaria parasite genomes!
There were two clear peaks. One was at 40% GC, similar to human genomes. But the other was at 20%, typical of malaria parasite genomes!
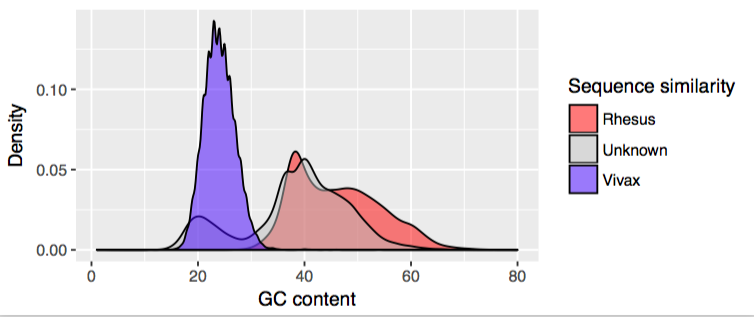
When I BLASTed each contig against related monkey and malaria species, it became pretty clear that almost all of the sequence in this peak at 20% was reminiscent of a malaria-like parasite.
The metadata said that the genome had been generated from the whole blood of a wild monkey. It seemed plausible that a bloodborne parasite genome might have been serendipitously captured as part of the host sequencing project.
But what parasite? A quick look at the literature suggested that Plasmodium infection was unlikely in this host. But a relative, Hepatocystis, was known to infect colobus monkeys.
Hepatocystis is related to the malaria parasites, but has crucial differences in its lifecycle.
Hepatocystis is related to the malaria parasites, but has crucial differences in its lifecycle.
It is transmitted not by mosquitoes, but by biting midges, and replicates only in the liver, not in the blood. (Sidebar: we owe both these insights to a multi-decade quest:)
No substantial Heptocystis genomic sequence has previously been described. But sequences for a handful of genes were in GenBank. Comparing the clp3 gene in the monkey assembly with its Hepatocystis counterpart confirmed that this monkey had been infected with Hepatocystis.
I did some preliminary work to annotate all of the Hepatocystis contigs with COMPANION, but ultimately decided that this was probably a job best done with the help of professionals, and also wanted to check it wasn& #39;t already work in progress. https://twitter.com/theosanderson/status/981971383923044352">https://twitter.com/theosande...
I wrote it all up in a blog post http://theo.io/blog/2018/04/23/how-i-stumbled-upon-a-novel-genome-for-a-malaria-like-parasite-of-apes/">https://theo.io/blog/2018... ...
And fortunately that exactly the right team came across it. @adamjamesreid decided to get in touch with @noahdsimons and his colleagues to discuss a collaboration, with Eerik leading the creation and analysis of a draft genome from a new assembly.
Uli lent all of the expertise she applies to the curation and annotation of the Plasmodium genomes that our community relies on to this new Hepatocystis sequence.
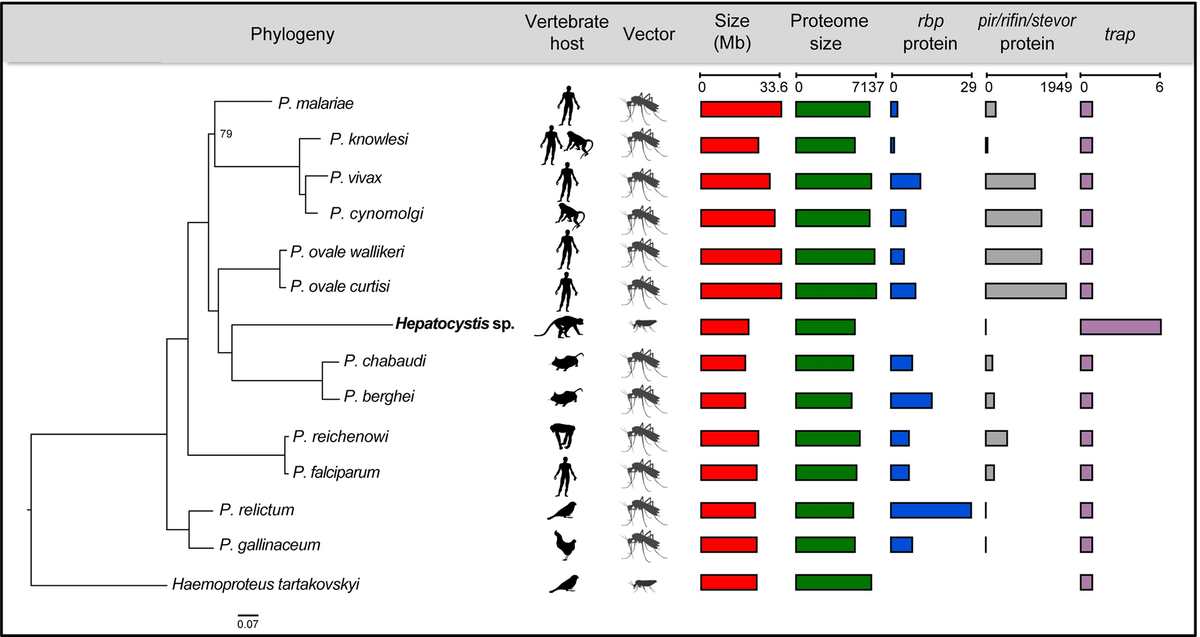
Our new genome is 19.95 Mb with 5,341 genes. Phylogenetic analysis of this genome confirms that Hepatocystis is nestled right within the Plasmodium genus.
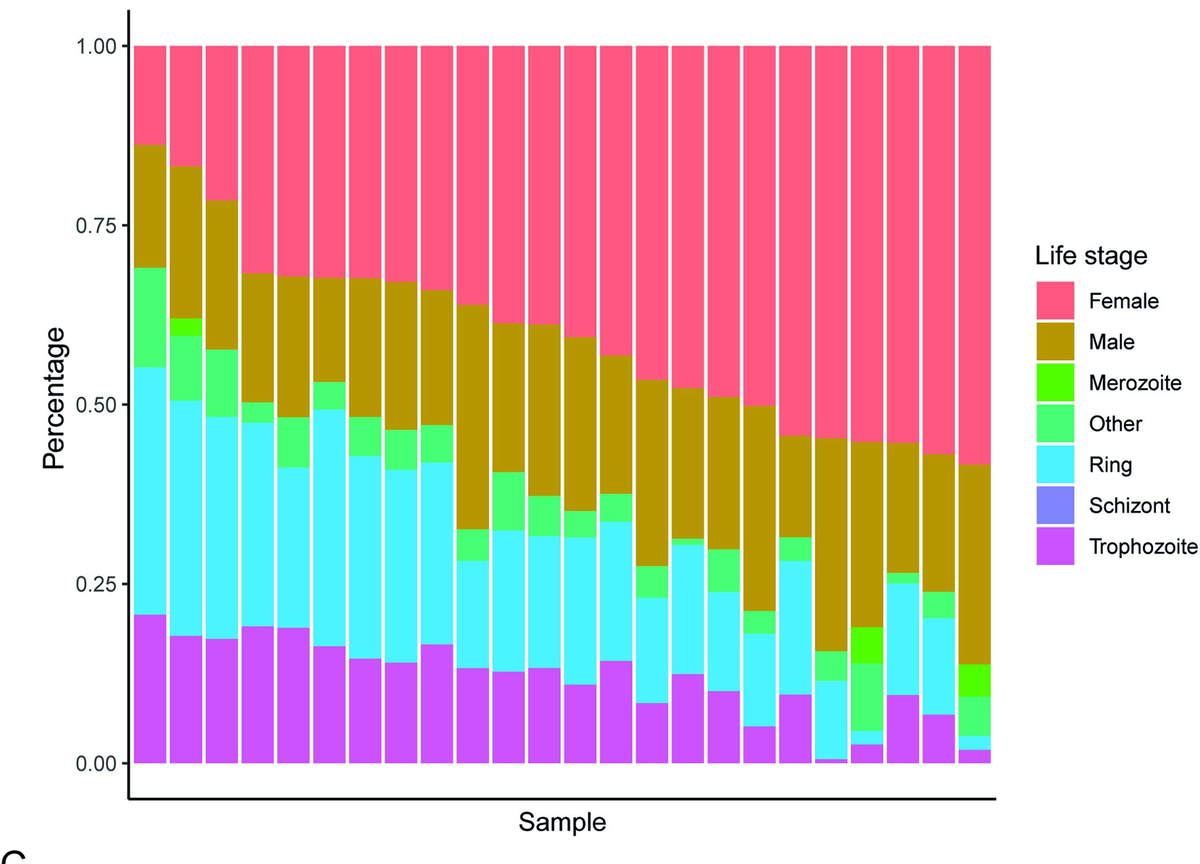
By analysing RNA-Seq data generated from this same population of monkeys by @noahdimons and colleagues, Eerik and Adam were able to shed light on the different lifestages circulating in the blood of these monkeys.
We deconvoluted the bulk RNA signal using single cell data from the Malaria Cell Atlas, and found that Hepatocystis the schizont expression that signals proliferation in the blood is absent in Hepatocystis.
We also picked up a genomic signal for this change in the lifecycle, with genes lost in Hepatocystis compared to Plasmodium enriched for schizont expression.
Indeed the Hepatocystis does not seem to encode a single RBP-family gene, as opposed to the many copies of this family of red-blood-cell binding proteins found in malaria parasites.
There& #39;s more. Evidence of a bunch of rapid evolution in some genes that might relate to adapting to a new insect host, and analysis of sex ratio from RNA-seq. Do check out the paper! #abstract1">https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1008717 #abstract1">https://journals.plos.org/plospatho...
It& #39;s been great to be part of a team across continents and disciplines pulling this together – thanks all!
Garnham spent 30 years on-and-off in his quest to understand Hepatocystis biology. I think he might be a bit galled to have learnt that today one could accidentally uncover its genome on the internet. But I like to think he& #39;d have taken it in good humour.

 Read on Twitter
Read on Twitter