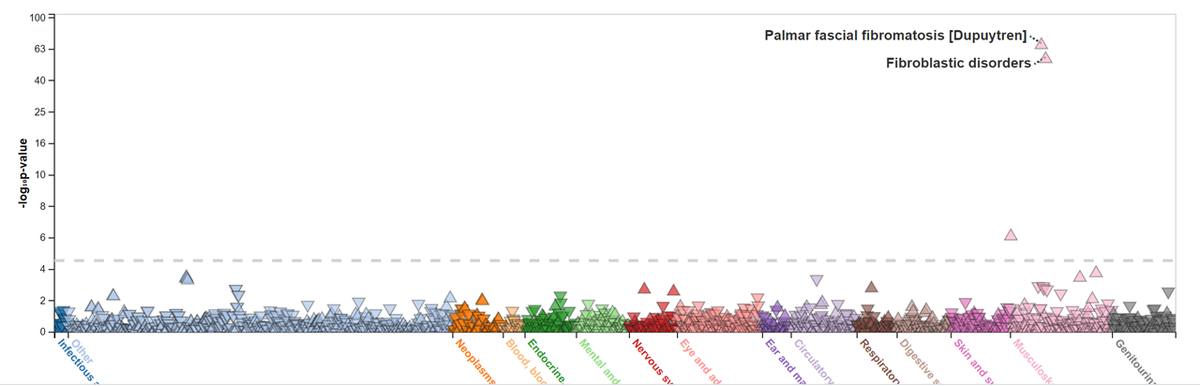
The first step in mapping a SNP to a gene is to understand what was measured. "Fibroblastic disorders" is a little vague, but going to the (very nice) PheWeb server we can better understand the trait (1/n) https://twitter.com/SbotGwa/status/1289545548660445184">https://twitter.com/SbotGwa/s...
It& #39;s always useful to look at the strongest association at a SNP of interest.
In this case, the strongest p-value is for Dupuytren& #39;s contracture, a specific form of "fibroblastic disorders"
https://r3.finngen.fi/variant/22-45971264-G-A">https://r3.finngen.fi/variant/2...
In this case, the strongest p-value is for Dupuytren& #39;s contracture, a specific form of "fibroblastic disorders"
https://r3.finngen.fi/variant/22-45971264-G-A">https://r3.finngen.fi/variant/2...
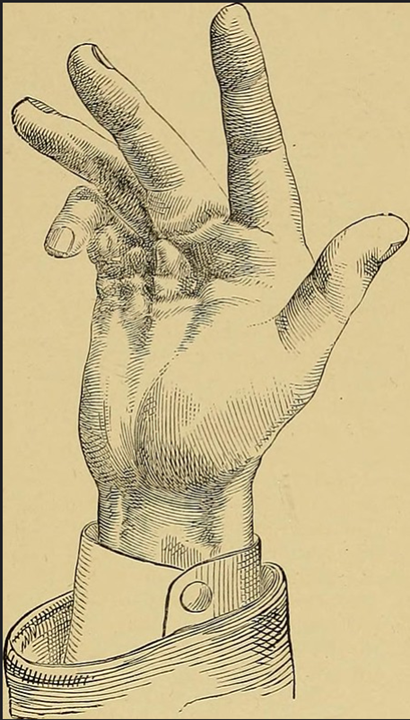
(3/n) Dupuytren& #39;s results from the formation of fibrous scar tissue around the tendons in the hand which forces the fingers to close:
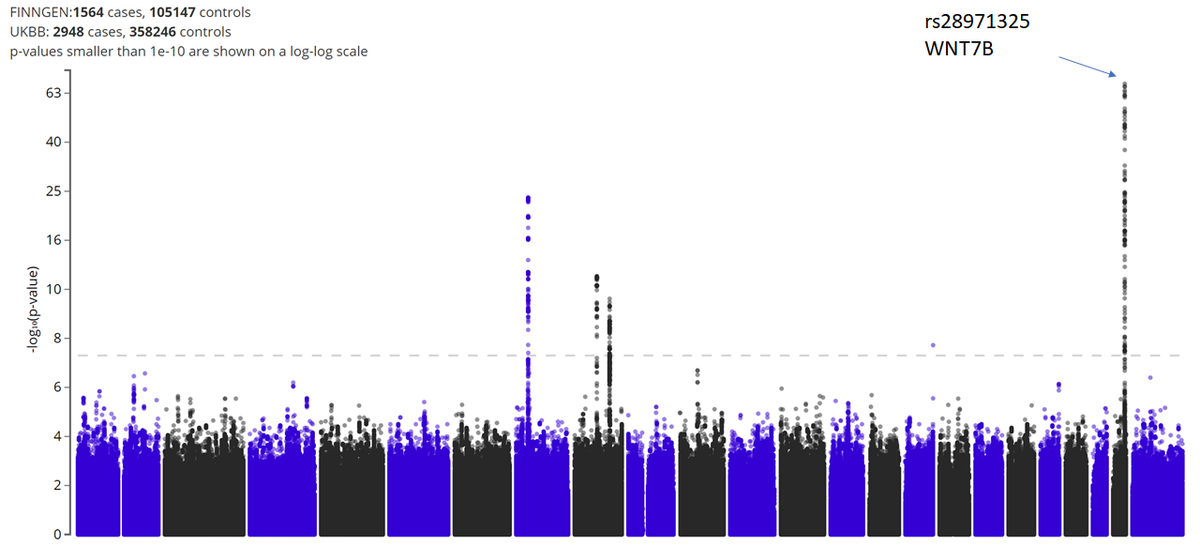
(4/n) now we can look at the FInnGen Dupuytren& #39;s GWAS http://r3.finngen.fi/pheno/M13_DUPUTRYEN">https://r3.finngen.fi/pheno/M13... which has 4 solid signals.
The closest gene isn& #39;t always the right gene, but it& #39;s a reasonable place to start.
The closest gene isn& #39;t always the right gene, but it& #39;s a reasonable place to start.
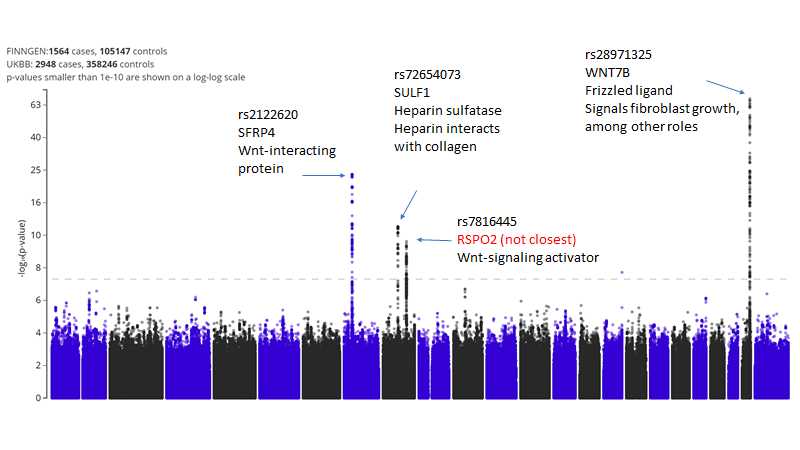
(5/n) WNT7B belongs to the WNT family of signaling molecules which initiate tissue and organ formation programs in multiple systems: https://en.wikipedia.org/wiki/Wnt_signaling_pathway">https://en.wikipedia.org/wiki/Wnt_...
(6/n) In fact as we review the top 4 hits most of them relate to Wnt signaling.
Also, at 3 of 4 loci the most likely causal gene is in fact the closest gene (75%)
Also, at 3 of 4 loci the most likely causal gene is in fact the closest gene (75%)
(7/n) If you like this, the strongest GWAS I can find on Dupuytren& #39;s is in this 2017 paper: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5591021/@EleZeggini">https://www.ncbi.nlm.nih.gov/pmc/artic...

 Read on Twitter
Read on Twitter