The #EtchACell #Preprint is out!  https://abs.twimg.com/emoji/v2/... draggable="false" alt="🎉" title="Partyknaller" aria-label="Emoji: Partyknaller">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🎉" title="Partyknaller" aria-label="Emoji: Partyknaller"> https://abs.twimg.com/emoji/v2/... draggable="false" alt="🎉" title="Partyknaller" aria-label="Emoji: Partyknaller">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🎉" title="Partyknaller" aria-label="Emoji: Partyknaller"> https://abs.twimg.com/emoji/v2/... draggable="false" alt="🎉" title="Partyknaller" aria-label="Emoji: Partyknaller">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🎉" title="Partyknaller" aria-label="Emoji: Partyknaller">
Includes contributions from thousands of volunteers from all around the world on our #CitizenScience project hosted on @the_zooniverse, analysing data from the @EM_STP team @TheCrick.
Thread with some results incoming, stay tuned! https://twitter.com/EtchACell/status/1288756652884336640">https://twitter.com/EtchACell...
Includes contributions from thousands of volunteers from all around the world on our #CitizenScience project hosted on @the_zooniverse, analysing data from the @EM_STP team @TheCrick.
Thread with some results incoming, stay tuned! https://twitter.com/EtchACell/status/1288756652884336640">https://twitter.com/EtchACell...
The problem: many microscopes produce datasets in the giga- to terabyte regime. Extracting meaning from these datasets is HARD!
E.g. 3D #ElectronMicroscopy methods like #SBFSEM and #FIBSEM produce data at 100s of GB per day
Movie: @GatanMicroscopy 3View https://www.youtube.com/watch?v=PWbLA7WMd34">https://www.youtube.com/watch...
E.g. 3D #ElectronMicroscopy methods like #SBFSEM and #FIBSEM produce data at 100s of GB per day
Movie: @GatanMicroscopy 3View https://www.youtube.com/watch?v=PWbLA7WMd34">https://www.youtube.com/watch...
We& #39;ve uploaded some benchmark datasets to the @EMDB_EMPIAR repository, why not download them and have a play! (You can read and convert the @GatanMicroscopy .DM4 files with @bioformats in @FijiSc)
E.g. this one used in the preprint: https://www.ebi.ac.uk/pdbe/emdb/empiar/entry/10094/">https://www.ebi.ac.uk/pdbe/emdb...
E.g. this one used in the preprint: https://www.ebi.ac.uk/pdbe/emdb/empiar/entry/10094/">https://www.ebi.ac.uk/pdbe/emdb...
This is a sequence of 518 slices with 8192x8192 pixels, covering a FOV of 82x82 microns and a depth of 26 microns (10x10x50nm voxel size). The sample is a pellet of #HeLa cells, embedded in resin, imaged by all-round EM legend Chris Peddie
Movie: scrolling through volume
Movie: scrolling through volume
The gold standard #BioImageAnalysis method for extracting information from this kind of data is manual #Segmentation, as demonstrated here by another @EM_STP team legend, Anne Weston (if you get a chance, check out some of Anne& #39;s images here: https://wellcomecollection.org/works?query=%22Anne%20Weston,%20Francis%20Crick%20Institute%22)">https://wellcomecollection.org/works...
This is a very accurate method, since an expert can make use of knowledge they have accumulated over their career. BUT, it is really time-consuming and  https://abs.twimg.com/emoji/v2/... draggable="false" alt="🤫" title="Schh!-Gesicht" aria-label="Emoji: Schh!-Gesicht">*whispers* experts don& #39;t always agree with each other!
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🤫" title="Schh!-Gesicht" aria-label="Emoji: Schh!-Gesicht">*whispers* experts don& #39;t always agree with each other!  https://abs.twimg.com/emoji/v2/... draggable="false" alt="😱" title="Vor Angst schreiendes Gesicht" aria-label="Emoji: Vor Angst schreiendes Gesicht">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="😱" title="Vor Angst schreiendes Gesicht" aria-label="Emoji: Vor Angst schreiendes Gesicht">
So we need a much faster method that produces results that are as objective as possible. So, call in the robots, right?  https://abs.twimg.com/emoji/v2/... draggable="false" alt="🤖" title="Robotergesicht" aria-label="Emoji: Robotergesicht">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🤖" title="Robotergesicht" aria-label="Emoji: Robotergesicht"> https://abs.twimg.com/emoji/v2/... draggable="false" alt="🤖" title="Robotergesicht" aria-label="Emoji: Robotergesicht">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🤖" title="Robotergesicht" aria-label="Emoji: Robotergesicht"> https://abs.twimg.com/emoji/v2/... draggable="false" alt="🤖" title="Robotergesicht" aria-label="Emoji: Robotergesicht"> #MachineLearning methods, particularly #DeepLearning, have taken #BioImageAnalysis by storm, producing amazing results in many applications...
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🤖" title="Robotergesicht" aria-label="Emoji: Robotergesicht"> #MachineLearning methods, particularly #DeepLearning, have taken #BioImageAnalysis by storm, producing amazing results in many applications...
...however, if you don& #39;t prepare your robots sufficiently well for the task, bad things can happen! You need to make sure that your #MachineLearning has been well-trained to deal with the inputs you& #39;re throwing at it! https://www.youtube.com/watch?v=PtiBe_kvAUk">https://www.youtube.com/watch...
So the problem comes back to gathering lots of training data to pass to the #MachineLearning system, like a teacher giving their students mock exams, with known answers, in preparation for their final exams. The problem is, there aren& #39;t enough experts to get all the data we need!
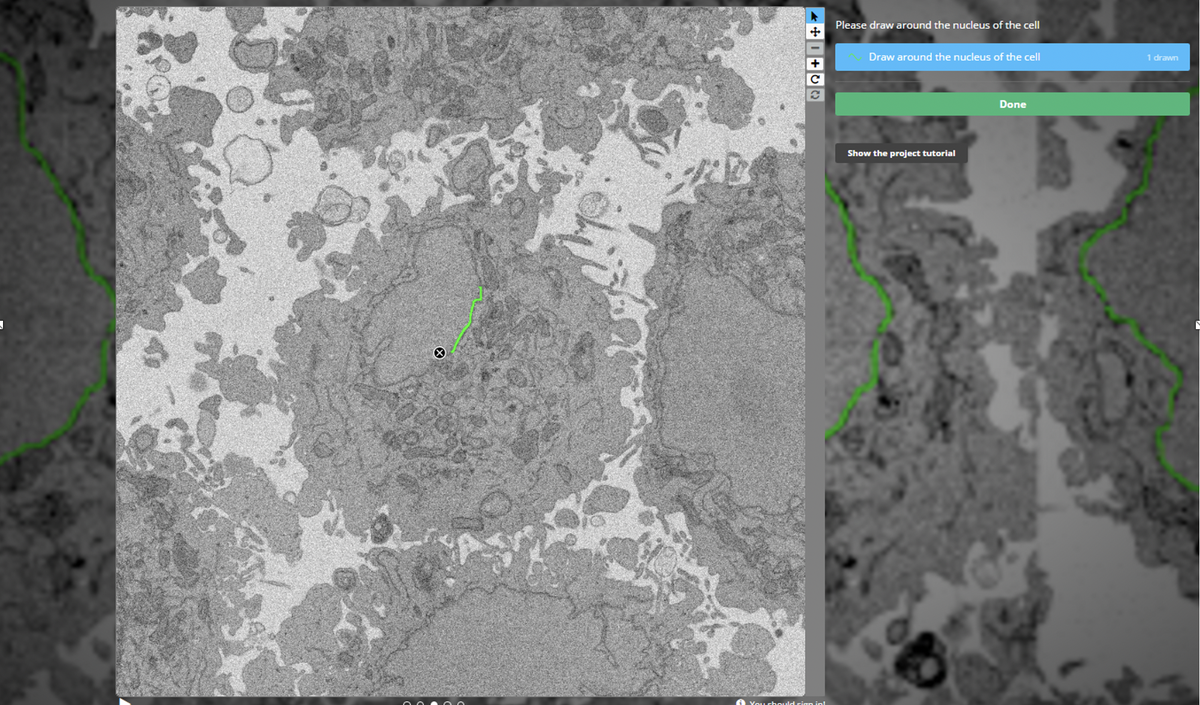
#Crowdsourcing of non-expert annotations has helped in many fields, from studying galaxies ( @galaxyzoo) to ecology ( @snapserengeti). We set up a project: @EtchACell, on @the_zooniverse platform, where we asked people to annotate our images in a browser https://www.zooniverse.org/organizations/h-spiers/etch-a-cell-colouring-in-cells-for-science">https://www.zooniverse.org/organizat...
The volume was chopped up and each small slice was shown to many volunteers. As you might expect, most individuals on their own were not quite as good as an expert, and we also had some "interesting" graffiti! Here& #39;s an example of the multiple annotations on a single slice
Here the task was to trace around the nuclear envelope (we are now targeting other organelles in other projects). Adding them up, you can see most volunteers do pretty well, but not perfectly. To make use of "the wisdom of the crowd", we need to aggregate the data more cleverly!
We tried lots of methods for this, finally arriving at "Contour Regression by Interior Averages" (CRIA  https://abs.twimg.com/emoji/v2/... draggable="false" alt="🦙" title="Llama" aria-label="Emoji: Llama">) - work by Luke Nightingale and Joost de Folter from the brilliant Scientific Computing STP @TheCrick, finding the boundary where volunteers agree on inside vs outside the NE
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🦙" title="Llama" aria-label="Emoji: Llama">) - work by Luke Nightingale and Joost de Folter from the brilliant Scientific Computing STP @TheCrick, finding the boundary where volunteers agree on inside vs outside the NE
Doing this for every slice of the ROIs allows us to piece together the nuclear envelope in 3D for many cells, purely from non-expert volunteer annotations. Here are segmented NEs from the 18 cells in the project (some incomplete due to not being fully within the acquired volume)
These are already plenty good enough for analysis at a much larger scale than usually possible. However, we& #39;re greedy and want to go even bigger, so we sent for the robots! This data is ideal for training a #DeepLearning system, like the ubiquitous #UNet ( https://lmb.informatik.uni-freiburg.de/Publications/2019/FMBCAMBBR19/)">https://lmb.informatik.uni-freiburg.de/Publicati...
By playing with scaling and views of the raw data, we can run the inference step across all 3 orthogonal views, "Tri-Axis Prediction" (TAP), which fixes problems inherent to a particular view (e.g. membrane oriented in the imaging/slicing plane) - H/T again to Joost and Luke!
Here the x-prediction=red, y-prediction=green, z-prediction=blue. Overlap of x+y but not z predictions gives yellow; x+z(not y) gives magenta; y+z(not x) gives cyan. Now we can identify and label NE that is hard, even for an expert!
Here are 3 reconstructions of the same nuclear envelope segmented with the 3 different methods: Gold: expert; Green: CRIA aggregated #CitizenScience; Magenta: 3D U-Net Tri-Axis Prediction, trained on non-expert #CitizenScience annotations alone.
We& #39;re very lucky at @TheCrick to have some fantastic computational resources (inc. @nvidia V100 GPUs) that mean our training (4hrs) and inference (~1 min/cell) are extremely fast. The trained #CNN can also be run over much larger input images, so here are the full FOV results
Here& #39;s a 3D rendering of full volume segmentation (made with @FijiSc& #39;s 3D Viewer). Inference time <50min + some post-processing (connected components etc)
An unexpected result was the ability of the model to deal with mitotic cells - there are no assumptions of "closed-ness" of the contours, so broken fragments of NE are found fairly well (not perfectly yet, but there were none in training!) For example, this cell from the volume:
Of course the dream is to create a system that generalises across different situations as well as an expert. However, many computational pipelines fail when even innocuous changes are made to your setup
Thanks to the large amount of #CitizenScience training data we got from #EtchACell (4241 images of 4 megapixel crops from the full volume) we found the model already generalises remarkably well to other imaging conditions
For example, here& #39;s the TAP #UNet prediction for a similar sample, but imaged in a different microscope ( #FIBSEM) at different resolution (5nm). As we get further from the original training data, we& #39;ll most likely need to employ further steps like transfer-learning or fine-tuning
As well as helping us with our research, the #EtchACell project has been a very successful #SciComm tool - volunteers are involved and invested in the research process and can hopefully relate more to scientists, helping to improve trust in science!
Alongside volunteers working on their own computers at home, #EtchACell has been at several outreach events, including @McrSciFest with @CRUKManchester, @NHM_London lates and @TheCrick& #39;s #CraftAndGraft exhibition (where we even got a contribution from @SadiqKhan @MayorofLondon!)
Links for more information:
Preprint: https://doi.org/10.1101/2020.07.28.223024
Code:">https://doi.org/10.1101/2... https://github.com/FrancisCrickInstitute/Etch-a-Cell-Nuclear-Envelope
CitSci">https://github.com/FrancisCr... data; aggregated and predicted segmentations: https://www.ebi.ac.uk/biostudies/studies/S-BSST448
Full">https://www.ebi.ac.uk/biostudie... raw data: https://dx.doi.org/10.6019/EMPIAR-10094
Cropped">https://dx.doi.org/10.6019/E... ROIs: https://dx.doi.org/10.6019/EMPIAR-10478">https://dx.doi.org/10.6019/E... (not live just yet, but soon!)
Preprint: https://doi.org/10.1101/2020.07.28.223024
Code:">https://doi.org/10.1101/2... https://github.com/FrancisCrickInstitute/Etch-a-Cell-Nuclear-Envelope
CitSci">https://github.com/FrancisCr... data; aggregated and predicted segmentations: https://www.ebi.ac.uk/biostudies/studies/S-BSST448
Full">https://www.ebi.ac.uk/biostudie... raw data: https://dx.doi.org/10.6019/EMPIAR-10094
Cropped">https://dx.doi.org/10.6019/E... ROIs: https://dx.doi.org/10.6019/EMPIAR-10478">https://dx.doi.org/10.6019/E... (not live just yet, but soon!)

 Read on Twitter
Read on Twitter