Out Today: our @NatureImmunol News & Views “The active inner life of naïve T cells” https://www.nature.com/articles/s41590-020-0726-1">https://www.nature.com/articles/... on the awesome #tcell #proteomics of @RGeigerLab “Dynamics in protein translation sustaining T cell preparedness”
10 reasons why this paper is super cool! … a thread
1/12
10 reasons why this paper is super cool! … a thread
1/12
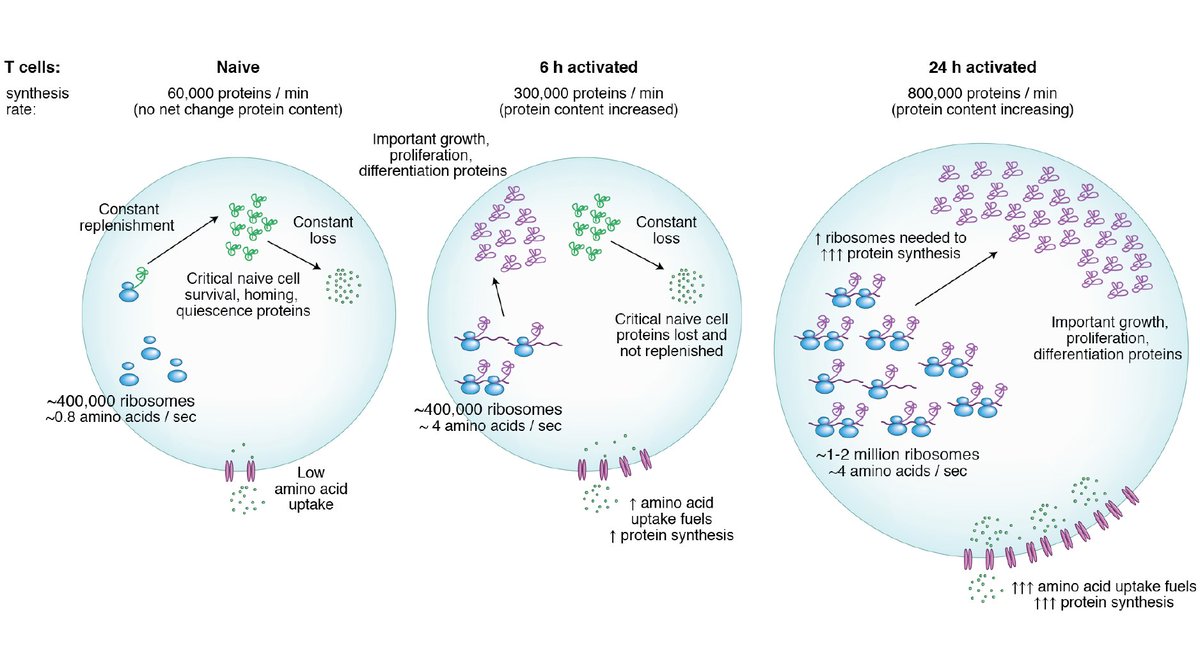
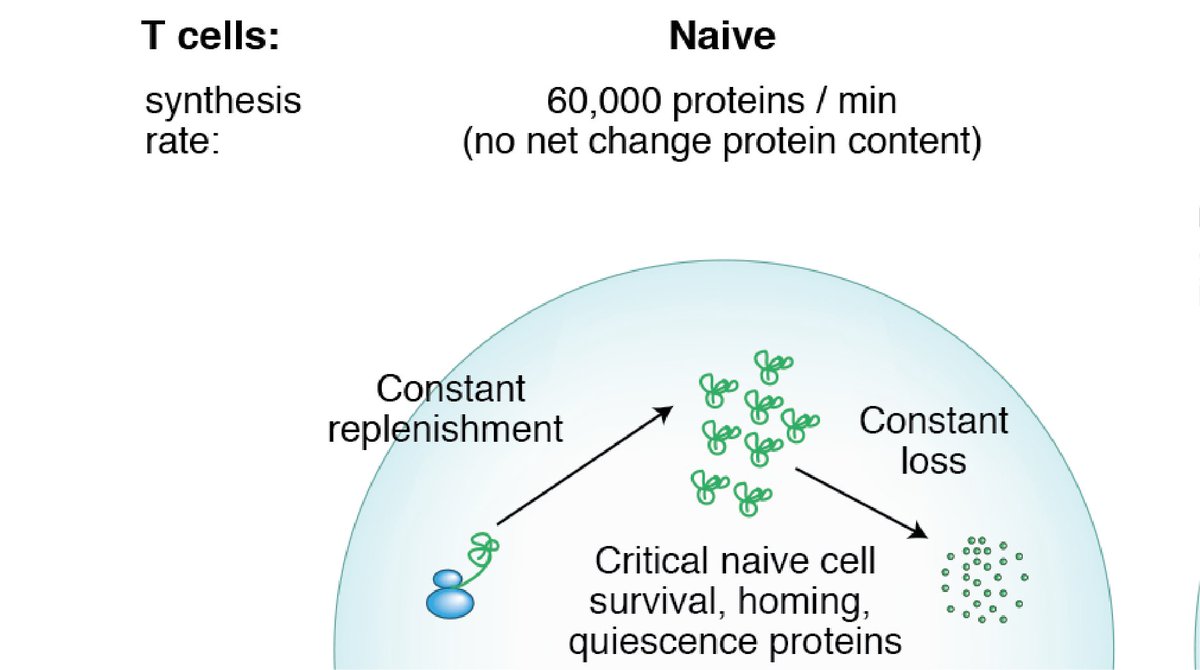
1) It busts the “inert” naïve T cell myth…. We all know that “naïve T cells remain in a ‘resting’ state until exposed to antigen”… but it’s not quite true! Naïve T cells are at least partially replenishing 19% of their protein species within the space of a day!
2) Many of the proteins that are turning over the fastest are the ones important for maintaining naïve T cell quiescence e.g. Tcf7, Foxo1, Ets1 (which had a half-life of just 53 minutes!) Naivety is an actively maintained state!
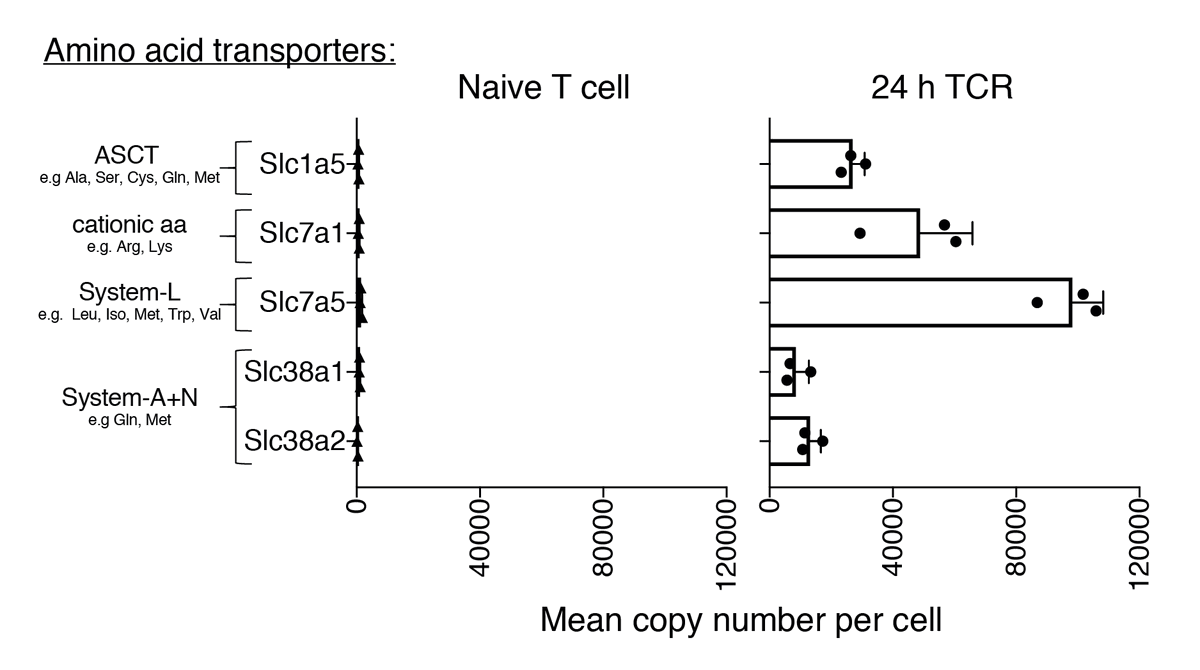
3) Protein turnover is measured by SILAC - heavy amino acids are transported into the cell and incorporated into new proteins. But naïve T cells have very low levels of amino acid transporters! See https://doi.org/10.7554/eLife.53725">https://doi.org/10.7554/e... Could protein turnover be even faster than measured here?
4) Naïve T cells turnover proteins by proteasomal degradation and #autophagy, an important process for survival and naivety. This study measured proteins that are still degraded when using a proteasome inhibitor – gives clues to the protein targets of autophagy in naïve T cells?
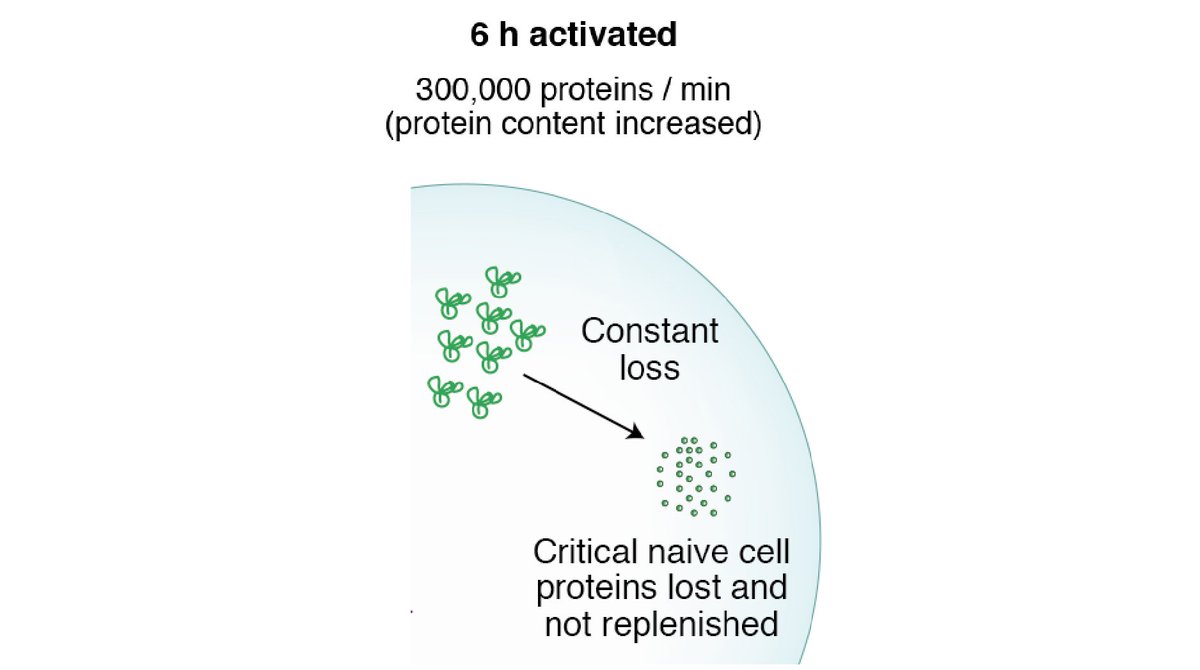
5) Many of the quickly turning over proteins in naïve T cells are lost during T cell activation – could this constant turnover be a ‘passive’ mechanism to ensure these proteins are destroyed when T cells activate?
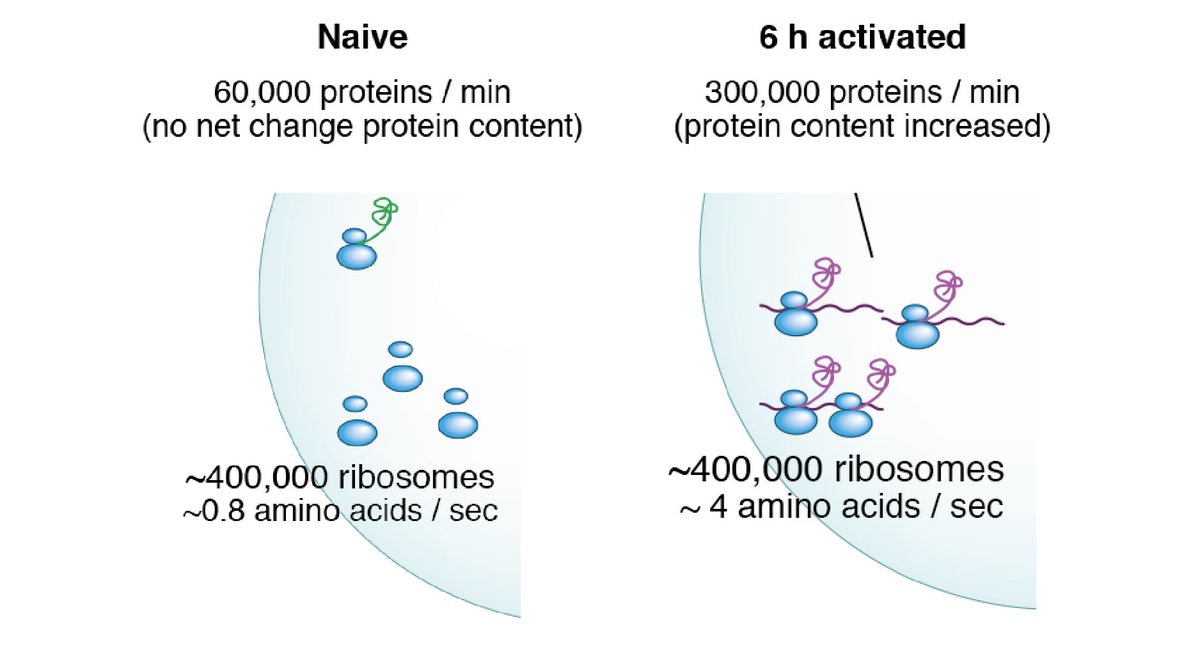
6) Calculations to work out proteins synthesised/min, # of ribosomes and therefore # aa/min/ribosome during T cell activation. Though ribosome number doesn’t change in the first 6 hours they go from idle to actively translating - This is how T cells can get going so quickly!
7) Calculations also show higher basal translation activity per ribosome in memory vs naive T cells - is this another way memory cells are poised for more rapid activation?
8) Clever estimates of mRNA copies / cell for each gene from RNAseq data. Since T cells increase their mRNA content as they activate use of values like TPM and FPKM can be misleading (mRNAs that don’t change expression may look reduced because other mRNAs are increasing)
9) Compares mRNA abundance vs protein translation rate to show which transcripts are repressed in naïve cells and poorly predict protein levels. A reminder the same mRNAs abundance can give a multiple orders of magnitude difference in translation rate! #thisiswhyweneedproteomics!
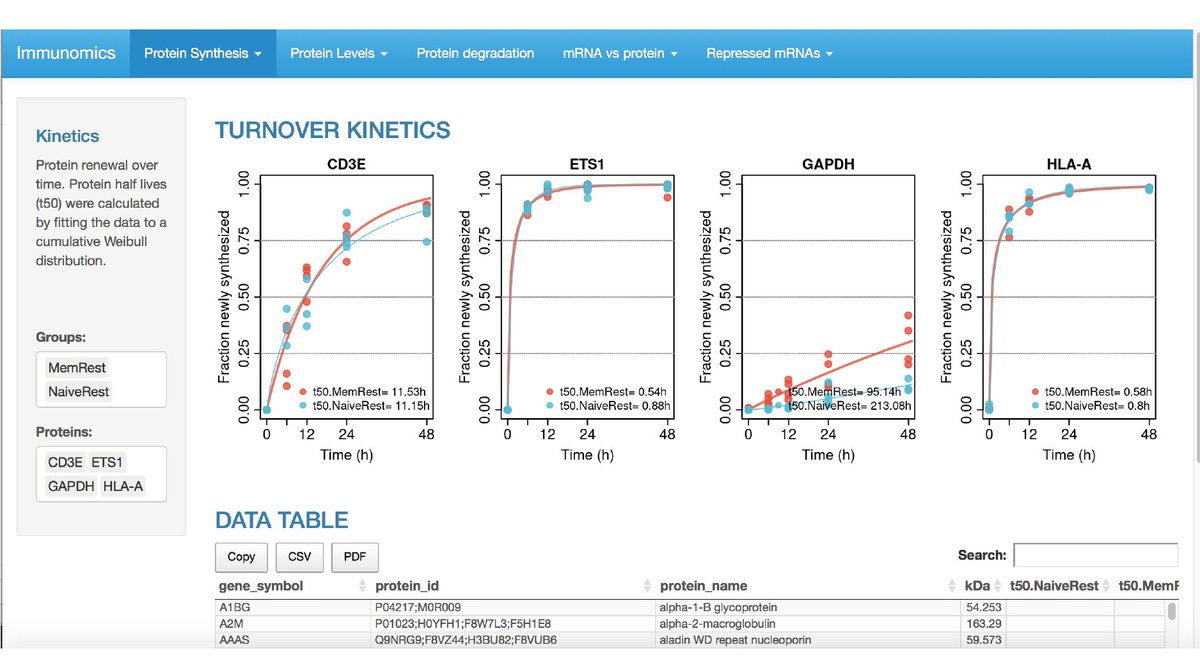
10) ... and my favourite things about this paper - The data is available on a super easy to use website: https://www.immunomics.ch"> https://www.immunomics.ch Hours of fun looking through the protein/mRNA dynamics await (and you don’t need a bioinformatics degree to be able to do it!)
And now that you’re all dying to read it here is a link to the primary research article: https://www.nature.com/articles/s41590-020-0714-5">https://www.nature.com/articles/... 12/12
Helps when I get the journal twitter handle correct!  https://abs.twimg.com/emoji/v2/... draggable="false" alt="😳" title="Errötetes Gesicht" aria-label="Emoji: Errötetes Gesicht"> ... Out Today: our @NatImmunol News & Views “The active inner life of naïve T cells” http://dx.doi.org/10.1038/s41590-020-0726-1">https://dx.doi.org/10.1038/s... on the awesome #tcell #proteomics of @RGeigerLab “Dynamics in protein translation sustaining T cell preparedness”
https://abs.twimg.com/emoji/v2/... draggable="false" alt="😳" title="Errötetes Gesicht" aria-label="Emoji: Errötetes Gesicht"> ... Out Today: our @NatImmunol News & Views “The active inner life of naïve T cells” http://dx.doi.org/10.1038/s41590-020-0726-1">https://dx.doi.org/10.1038/s... on the awesome #tcell #proteomics of @RGeigerLab “Dynamics in protein translation sustaining T cell preparedness”

 Read on Twitter
Read on Twitter