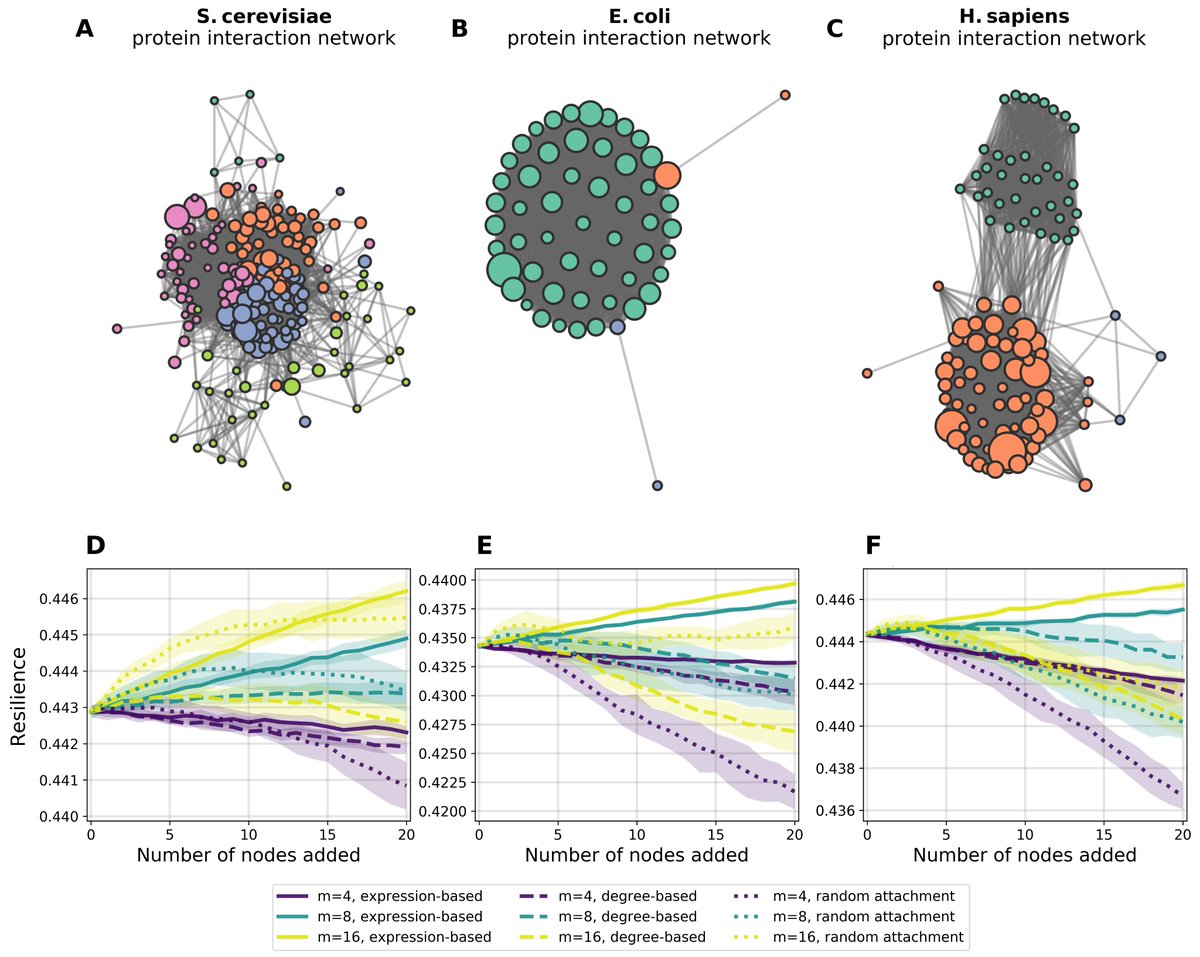
New preprint: When a new node (novelty) is added to a (protein) network, how (and why!) does that network& #39;s resilience change?
Work from the @sfiscience CSSS, led by @aprilkleppe w/ @lholmerx @kesm_ith @mmjohnEEB @anshuman2111 @laura_stolp @Ashley_Teufel
https://www.biorxiv.org/content/10.1101/2020.07.02.184325v1">https://www.biorxiv.org/content/1...
Work from the @sfiscience CSSS, led by @aprilkleppe w/ @lholmerx @kesm_ith @mmjohnEEB @anshuman2111 @laura_stolp @Ashley_Teufel
https://www.biorxiv.org/content/10.1101/2020.07.02.184325v1">https://www.biorxiv.org/content/1...
Some deets:
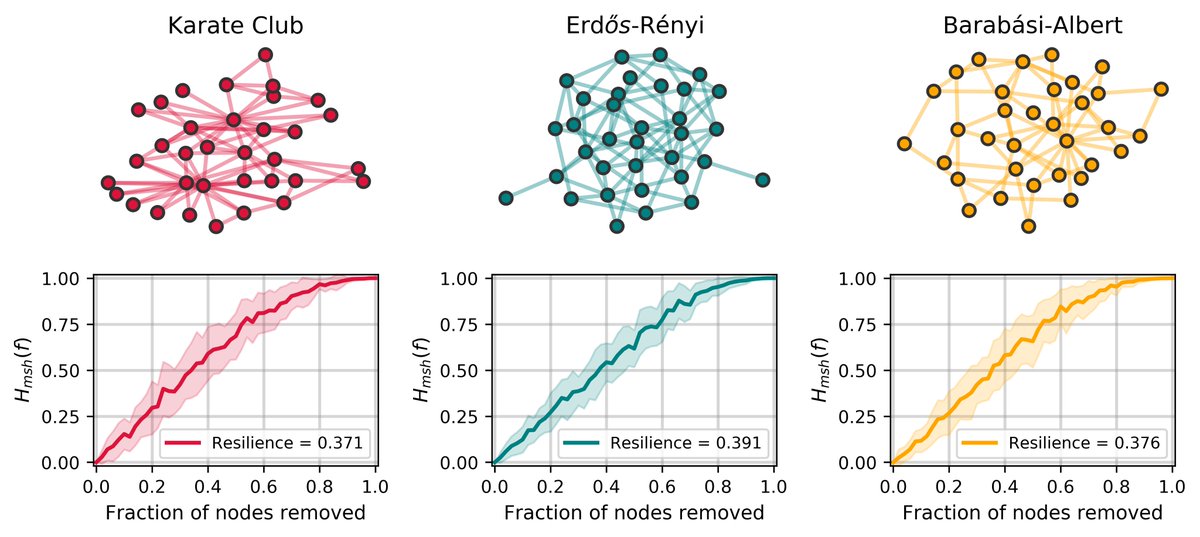
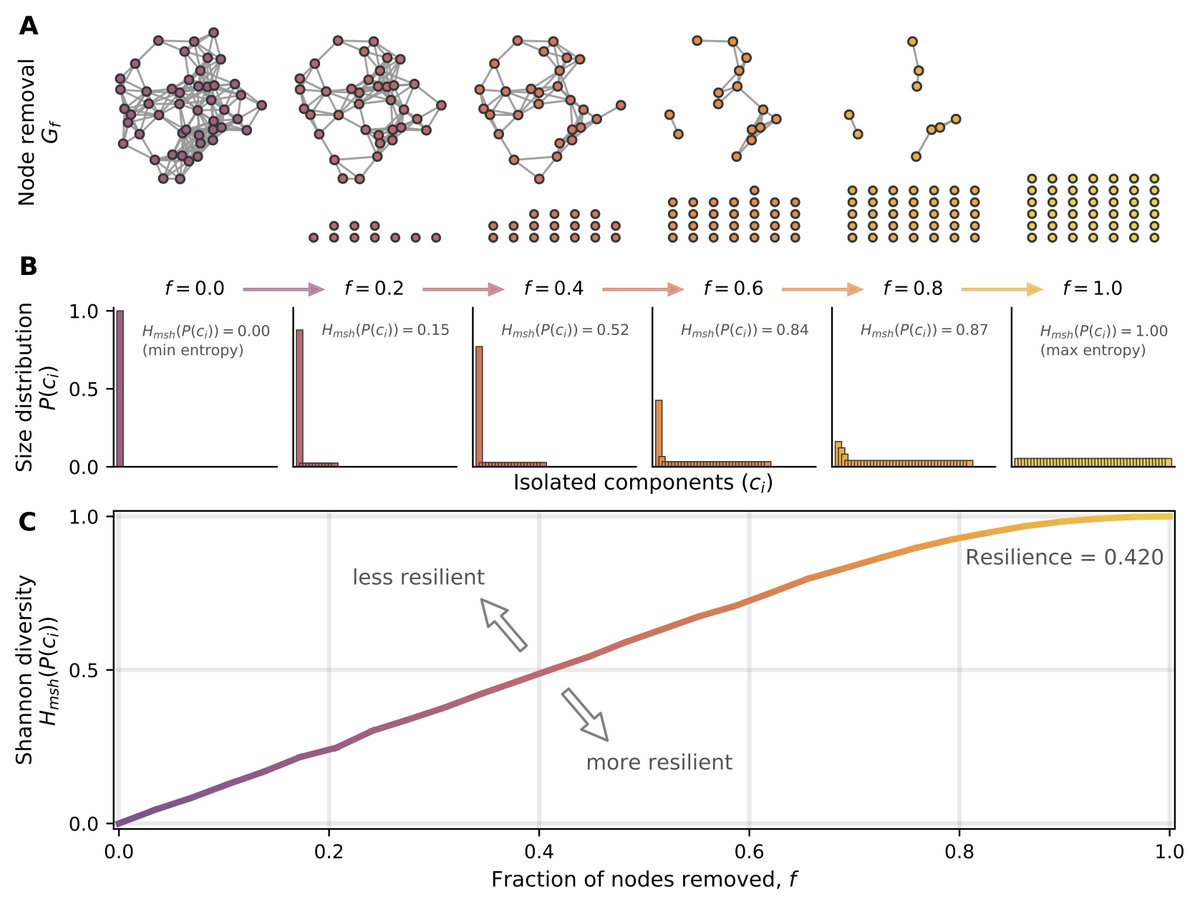
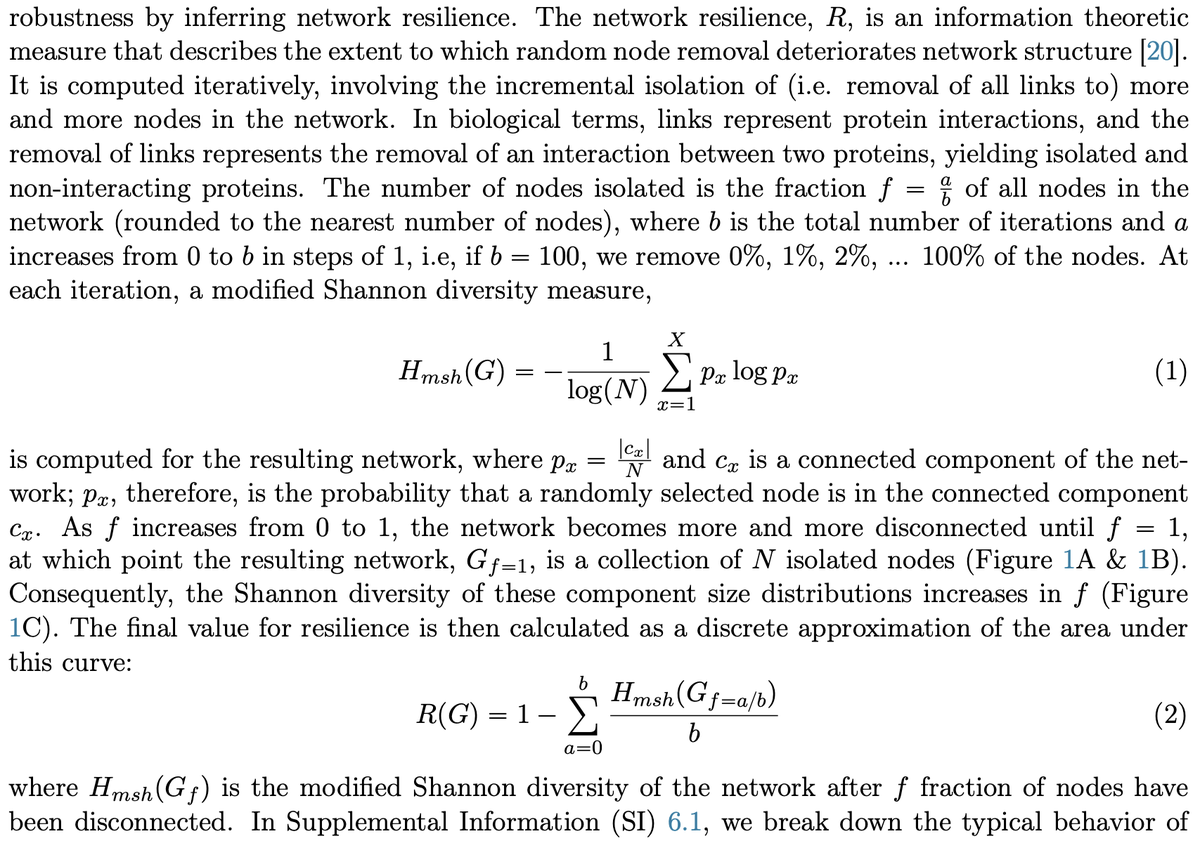
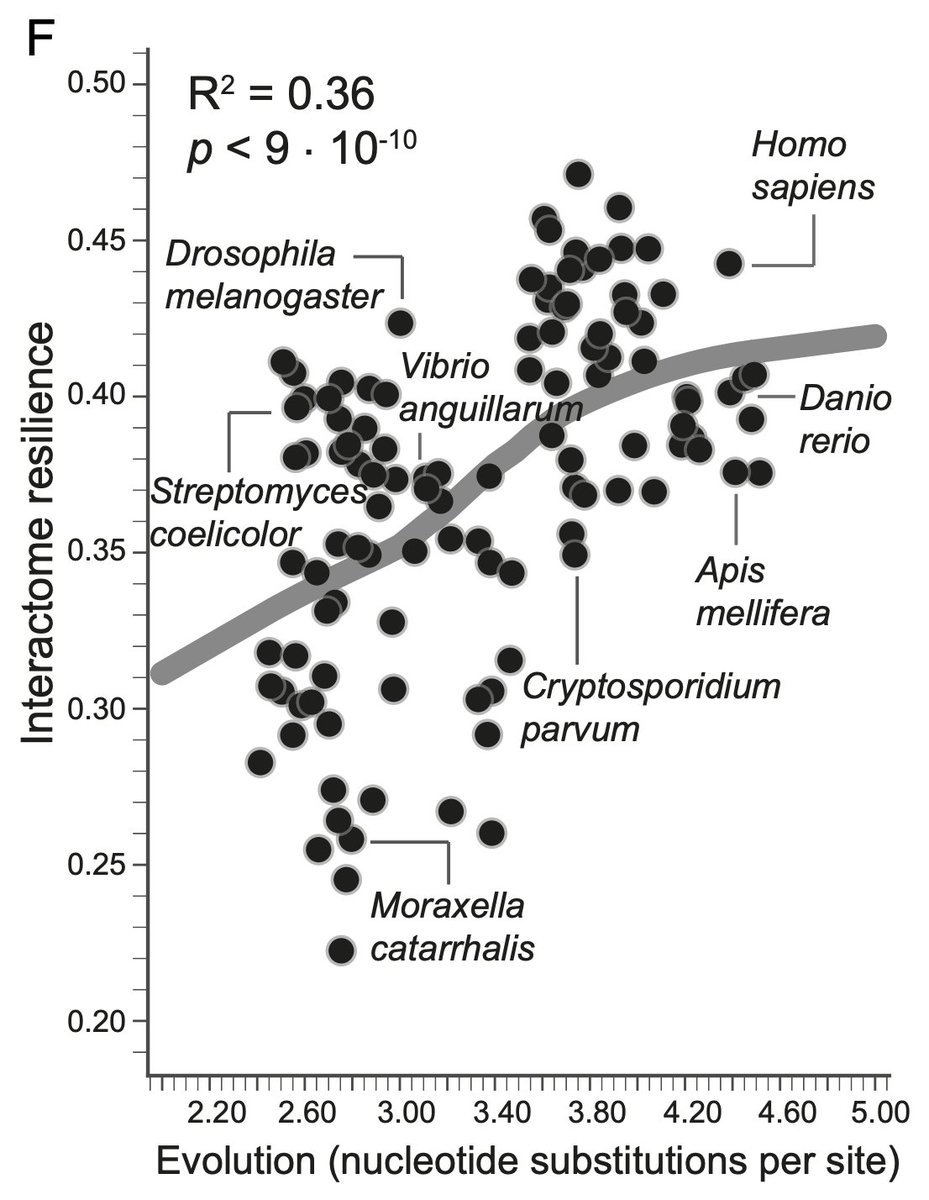
We got the idea of network "resilience" from a great paper ( https://www.pnas.org/content/116/10/4426)">https://www.pnas.org/content/1... by @marinkazitnik and co.
It& #39;s basically the entropy of the component sizes distrib as you iteratively disconnect more nodes
(sidenote this certainly could be called robustness) 1/
We got the idea of network "resilience" from a great paper ( https://www.pnas.org/content/116/10/4426)">https://www.pnas.org/content/1... by @marinkazitnik and co.
It& #39;s basically the entropy of the component sizes distrib as you iteratively disconnect more nodes
(sidenote this certainly could be called robustness) 1/
Their paper showed that different species& #39; protein networks can have a range of resilience values, which gave us an idea:
If resilience is defined by removing nodes, how does the resilience of protein networks change when adding new nodes?
A https://abs.twimg.com/emoji/v2/... draggable="false" alt="🔆" title="Symbol für starke Helligkeit" aria-label="Emoji: Symbol für starke Helligkeit">presilience
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🔆" title="Symbol für starke Helligkeit" aria-label="Emoji: Symbol für starke Helligkeit">presilience https://abs.twimg.com/emoji/v2/... draggable="false" alt="🔆" title="Symbol für starke Helligkeit" aria-label="Emoji: Symbol für starke Helligkeit"> if you will 2/
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🔆" title="Symbol für starke Helligkeit" aria-label="Emoji: Symbol für starke Helligkeit"> if you will 2/
If resilience is defined by removing nodes, how does the resilience of protein networks change when adding new nodes?
A
We& #39;re imagining evolutionary novelty as the introduction of something new into a system, which doesn& #39;t in turn destroy the organism. *Of course, it& #39;s not this simple.
But in our context, we& #39;re curious about the node attachment mechanisms that increase a network& #39;s resilience. 3/
But in our context, we& #39;re curious about the node attachment mechanisms that increase a network& #39;s resilience. 3/
So! We simulate a bunch of new nodes being added to a few different ribosomal protein networks under three different node-attachment mechanisms
- uniformly at random
- degree-based preferential attachment
- *gene-expression-based* pref. attach.
/4
- uniformly at random
- degree-based preferential attachment
- *gene-expression-based* pref. attach.
/4
I found this interesting: the presilience is highest when new nodes (proteins) preferentially attach to nodes already in the network *proportionally based on the nodes& #39; gene expression*
This suggests a potential relationship between gene expression & protein network structure /5
This suggests a potential relationship between gene expression & protein network structure /5
Then there& #39;s a whole fun SI section where we look at the resilience of different toy networks as we vary their parameters.
We also show that the maximum value for a network& #39;s resilience is 0.5 (previously described as 1.0)
/6
We also show that the maximum value for a network& #39;s resilience is 0.5 (previously described as 1.0)
/6
And some open code! https://github.com/jkbren/presilience
In">https://github.com/jkbren/pr... the repo, we actually go into a little more detail and toy examples in three tutorial-esque notebooks.
Also included the matplotlib code for making the figs. /7
In">https://github.com/jkbren/pr... the repo, we actually go into a little more detail and toy examples in three tutorial-esque notebooks.
Also included the matplotlib code for making the figs. /7
tl;dr - here& #39;s a pic that the 8 of us took in Santa Fe.
Getting to do science with your buddies is one of the best parts of being a scientist.
8/8
Getting to do science with your buddies is one of the best parts of being a scientist.
8/8

 Read on Twitter
Read on Twitter
 presiliencehttps://abs.twimg.com/emoji/v2/... draggable="false" alt="🔆" title="Symbol für starke Helligkeit" aria-label="Emoji: Symbol für starke Helligkeit"> if you will 2/" title="Their paper showed that different species& #39; protein networks can have a range of resilience values, which gave us an idea:If resilience is defined by removing nodes, how does the resilience of protein networks change when adding new nodes? A https://abs.twimg.com/emoji/v2/... draggable="false" alt="🔆" title="Symbol für starke Helligkeit" aria-label="Emoji: Symbol für starke Helligkeit">presiliencehttps://abs.twimg.com/emoji/v2/... draggable="false" alt="🔆" title="Symbol für starke Helligkeit" aria-label="Emoji: Symbol für starke Helligkeit"> if you will 2/" class="img-responsive" style="max-width:100%;"/>
presiliencehttps://abs.twimg.com/emoji/v2/... draggable="false" alt="🔆" title="Symbol für starke Helligkeit" aria-label="Emoji: Symbol für starke Helligkeit"> if you will 2/" title="Their paper showed that different species& #39; protein networks can have a range of resilience values, which gave us an idea:If resilience is defined by removing nodes, how does the resilience of protein networks change when adding new nodes? A https://abs.twimg.com/emoji/v2/... draggable="false" alt="🔆" title="Symbol für starke Helligkeit" aria-label="Emoji: Symbol für starke Helligkeit">presiliencehttps://abs.twimg.com/emoji/v2/... draggable="false" alt="🔆" title="Symbol für starke Helligkeit" aria-label="Emoji: Symbol für starke Helligkeit"> if you will 2/" class="img-responsive" style="max-width:100%;"/>