Our new study on the genetic basis of #mosquito resistance to #DENV is now out in @PLOSGenetics. Congratulations to @Dickson14Laura, @Drosophilista, Mathieu Gautier, @xealf8, Davy Jiolle, Christophe Paupy, @d_ayalag, Isabelle Conclois and @albinfont ! https://dx.plos.org/10.1371/journal.pgen.1008794">https://dx.plos.org/10.1371/j...
In many invertebrate host-pathogen systems, infection success depends on the specific pairing of host and pathogen genotypes. We previously reported such genotype-by-genotype (G x G) interactions between dengue virus (DENV) and its main mosquito vector Aedes aegypti. A thread 1/7
Host genes underlying G x G interactions have largely remained elusive so far. We combined a natural phenotype of DENV type-specific resistance in Ae. aegypti and the power of high-throughput sequencing to provide insights into the genetic architecture of G x G interactions. 2/7
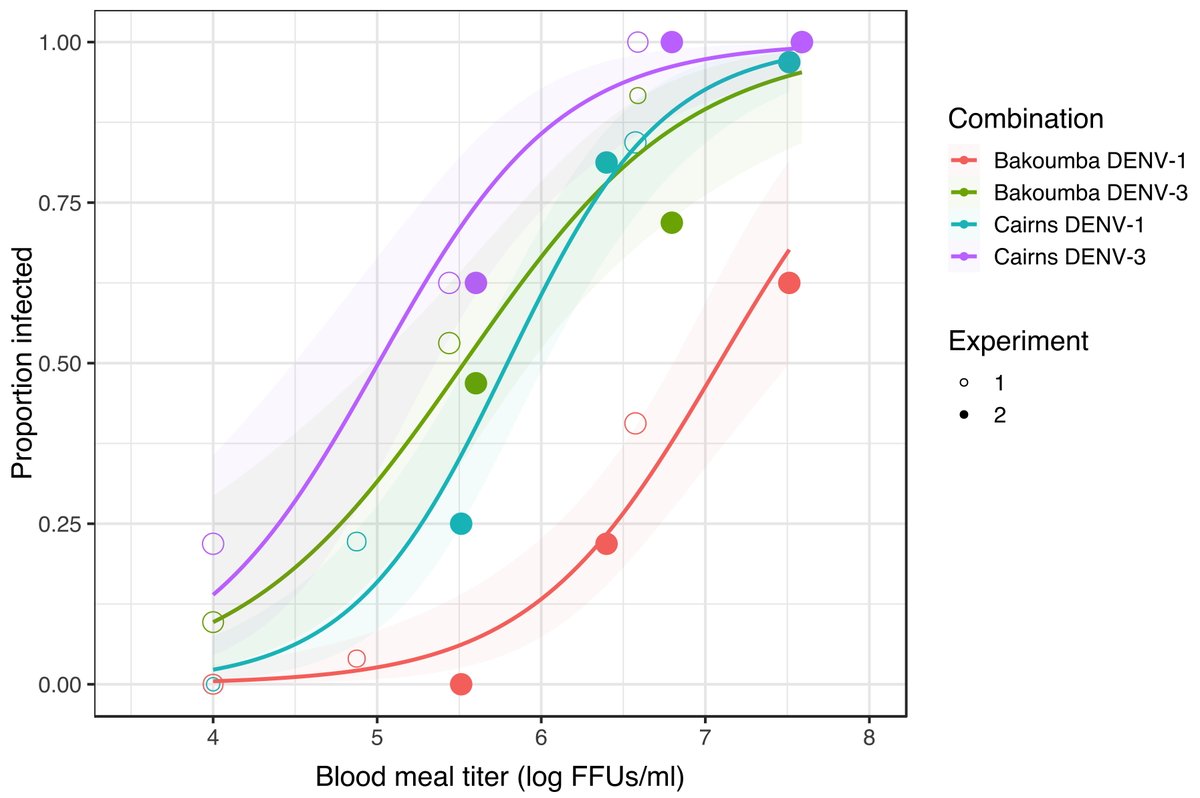
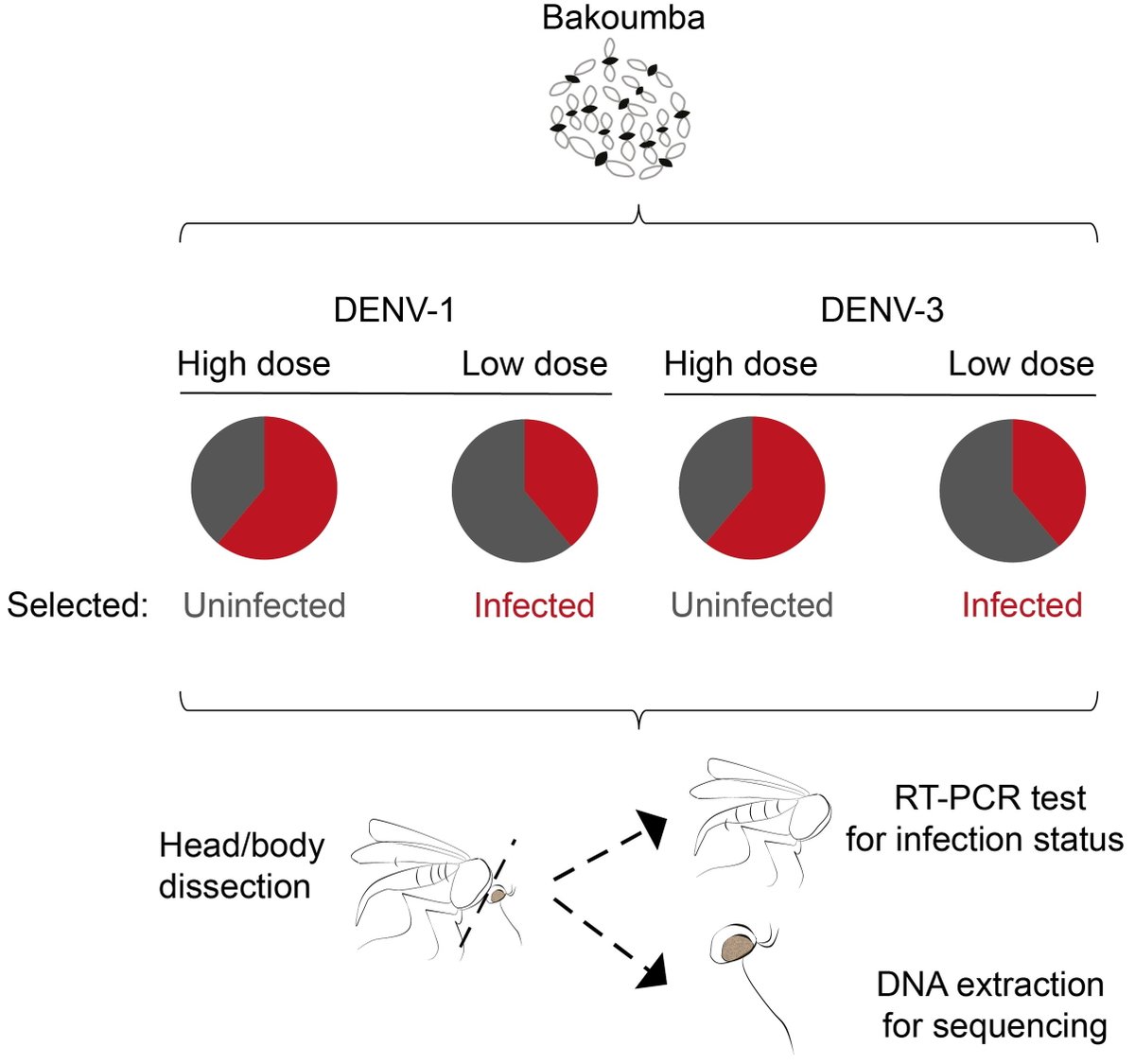
We previously established a lab colony from a wild Ae. aegypti population in Bakoumba, Gabon that is differentially susceptible to DENV types. In dose-response experiments, the Bakoumba population was strongly resistant to DENV-1 and only moderately resistant to DENV-3. 3/7
We compared the genetic basis of Bakoumba resistance to DENV-1 and DENV-3 by detecting allele frequency differences between replicate pools of individuals with contrasted phenotypes (resistant = uninfected at a high virus dose vs. susceptible = infected at a low virus dose). 4/7
To estimate and compare allele frequencies among the phenotypic pools, we performed exome sequencing as a cost-effective genotyping method targeting the exons of all protein-coding genes. 5/7
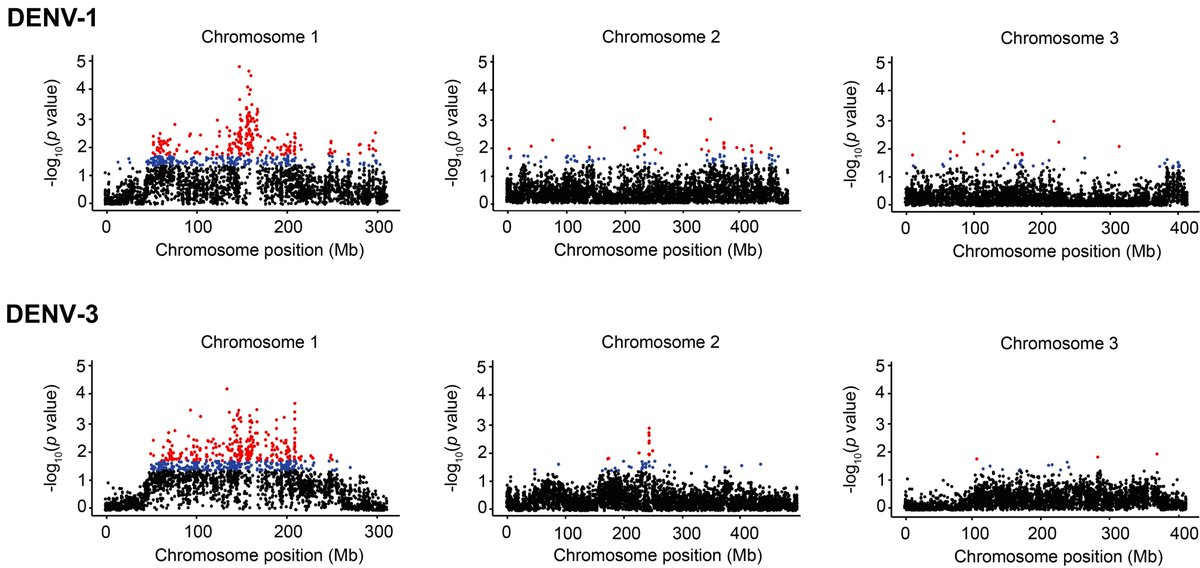
We computed gene scores reflecting the statistical significance of association with DENV-1 and/or DENV-3 infection. We found that largely distinct gene sets underlie specific resistance to DENV-1 and DENV-3 infection in this Ae. aegypti population. 6/7
Our findings indicate that different sets of host genes, rather than different variants of the same genes, confer pathogen-specific resistance in this system. This is an important step towards identification of underlying molecular mechanisms. 7/7

 Read on Twitter
Read on Twitter