Proud to announce our new manuscript from the @hilsomartin & @mehurles groups @sangerinstitute. We did a deep dive into the relationship between rare variant burden and reproductive success with the amazing data from @uk_biobank!
https://www.biorxiv.org/content/10.1101/2020.05.26.116111v1
Tweet">https://www.biorxiv.org/content/1... summary https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Down pointing backhand index" aria-label="Emoji: Down pointing backhand index">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Down pointing backhand index" aria-label="Emoji: Down pointing backhand index">
https://www.biorxiv.org/content/10.1101/2020.05.26.116111v1
Tweet">https://www.biorxiv.org/content/1... summary
This started as a rotation project led by the amazing Sanger PhD student @MDC_Neville supervised by @ksamocha, myself and our PI @mehurles. Following some cool initial findings, it "evolved" into what you see today.
As the flurry of #gnomAD papers has made obvious, some genes in the human genome are depleted for Protein Truncating Vars (PTVs). But only ~35% have an associated disease; why are genes without an associated disease depleted for PTVs? Could it be due to an effect on reproduction?
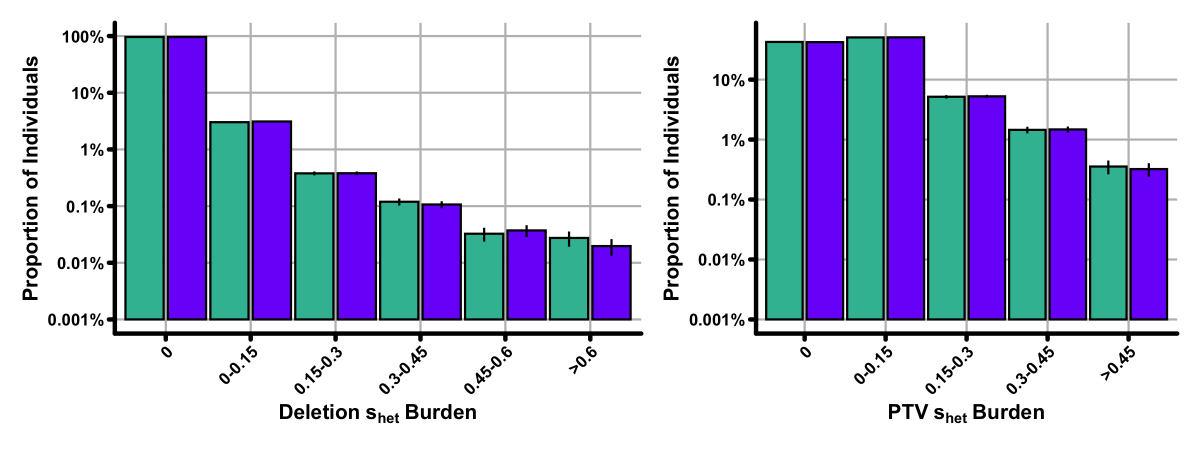
To investigate, we turned to the amazing @uk_biobank resource. For each participant we first created a "burden score" for genes disrupted by either a Deletion or PTV. ~1% of individuals have "high" burden (>0.15) as measured by Deletions, ~10% for PTVs, with no M/F bias.
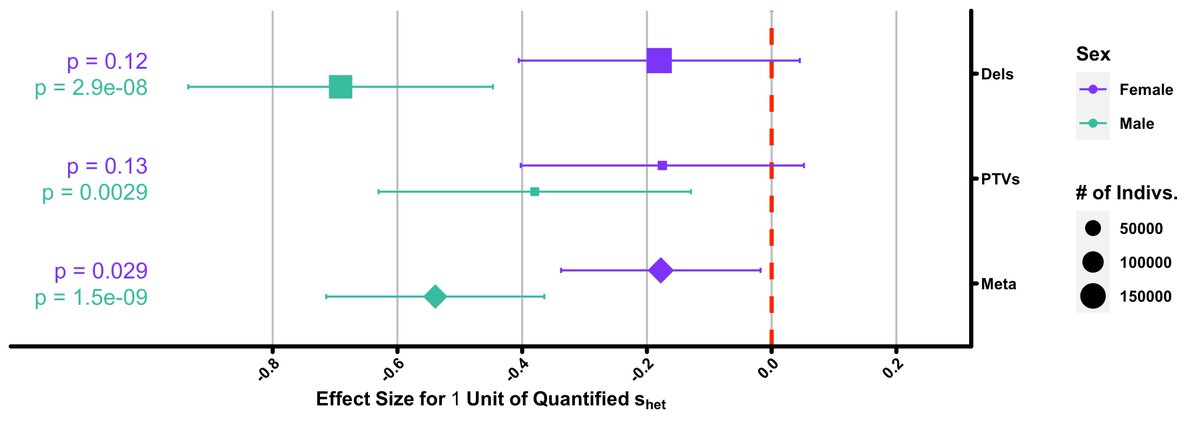
We then tested the association of
"# of children ~ s-het- burden"
and saw a striking sex stratified result:
"# of children ~ s-het- burden"
and saw a striking sex stratified result:
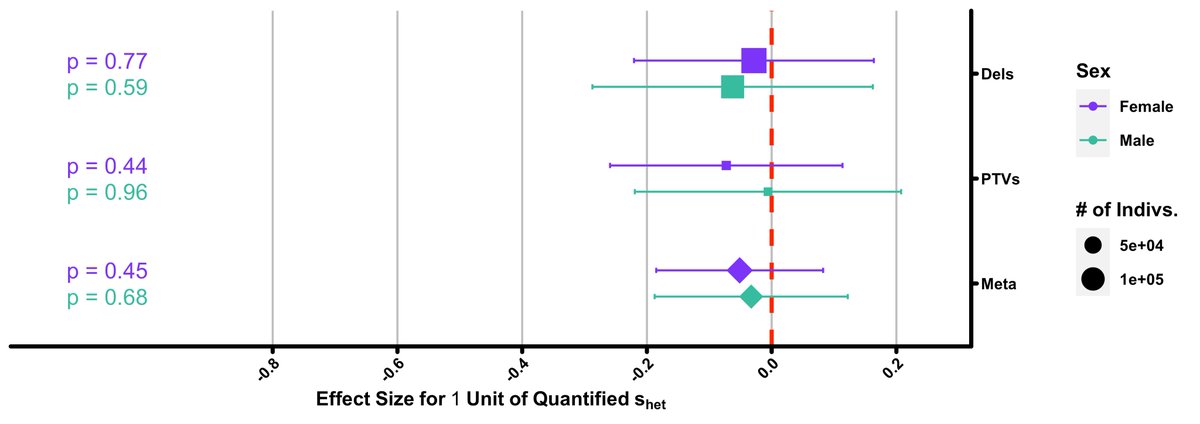
We next investigated if our observed result was due to an actual decrease in number of children or due to a decrease in having any children at all (ie childlessness). We found s-het- burden has no association with the former…
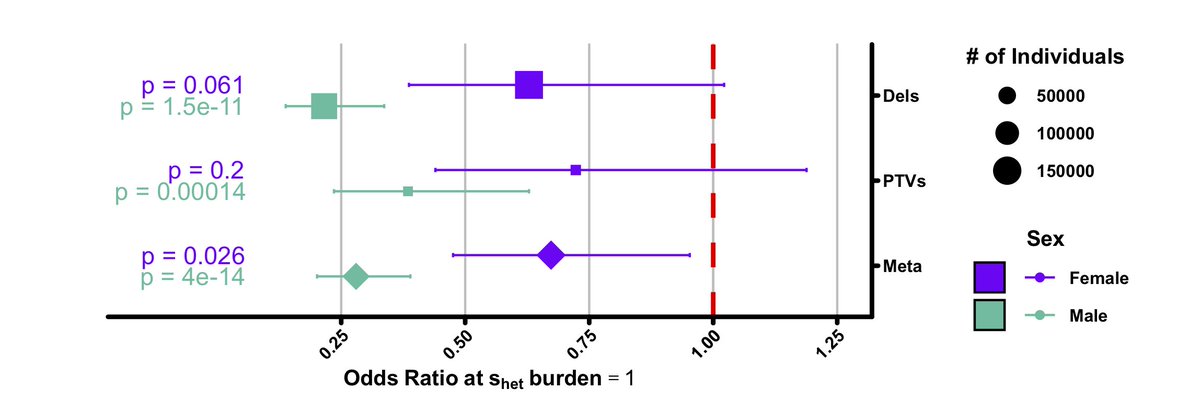
But we did find that s-het burden has a strong association with childlessness, in particular among males.
So how is our observed effect getting from:
Rare variant burden https://abs.twimg.com/emoji/v2/... draggable="false" alt="👉" title="Right pointing backhand index" aria-label="Emoji: Right pointing backhand index"> phenotype
https://abs.twimg.com/emoji/v2/... draggable="false" alt="👉" title="Right pointing backhand index" aria-label="Emoji: Right pointing backhand index"> phenotype  https://abs.twimg.com/emoji/v2/... draggable="false" alt="👉" title="Right pointing backhand index" aria-label="Emoji: Right pointing backhand index"> childlessness
https://abs.twimg.com/emoji/v2/... draggable="false" alt="👉" title="Right pointing backhand index" aria-label="Emoji: Right pointing backhand index"> childlessness
With a distinct male bias?
We investigated several hypotheses, including sociodemographic and more specific factors like loss of genes associated with male infertility.
Rare variant burden
With a distinct male bias?
We investigated several hypotheses, including sociodemographic and more specific factors like loss of genes associated with male infertility.
But…
“it’s probably not due to infertility” - us
And we also didn& #39;t find significant sex-bias for most sociodemographic traits like income, cognition, or education.
“it’s probably not due to infertility” - us
And we also didn& #39;t find significant sex-bias for most sociodemographic traits like income, cognition, or education.
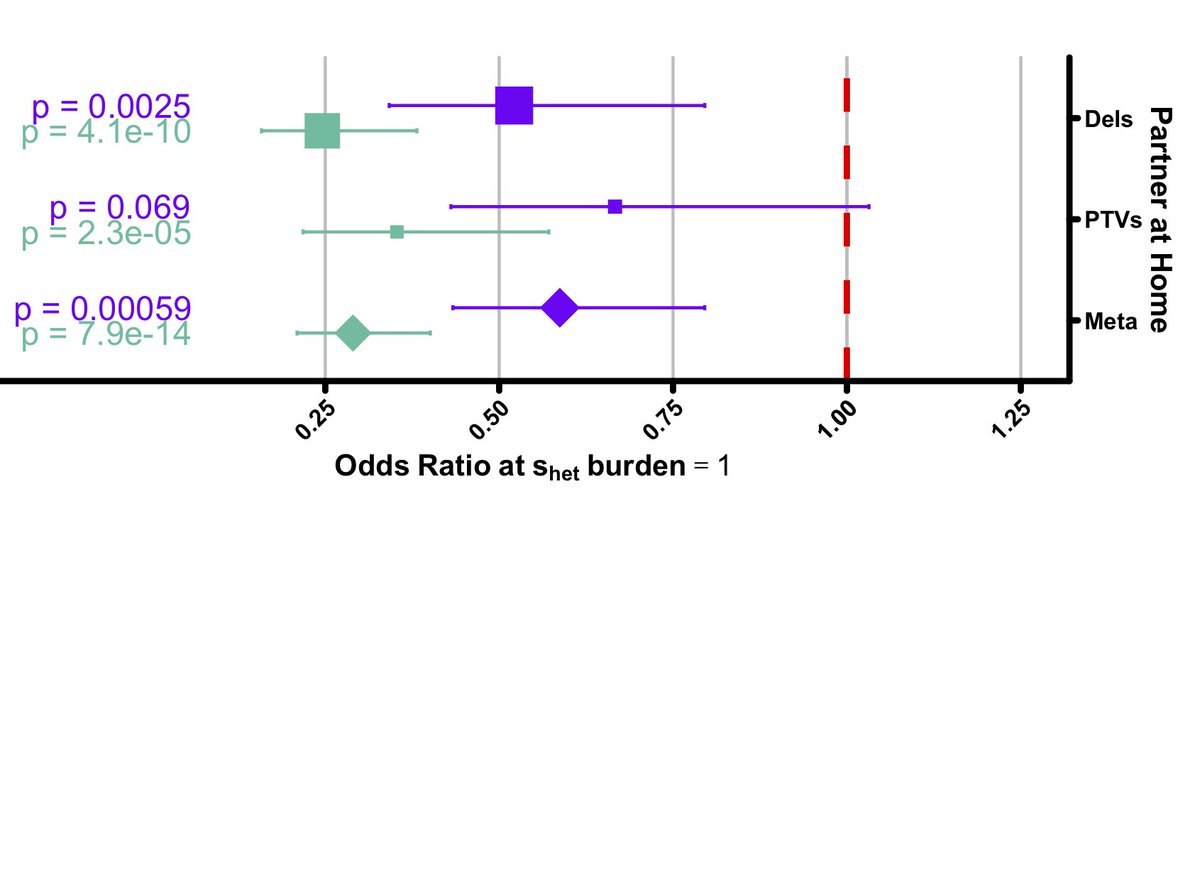
But what we did find was a strong male-specific bias in "currently living with a partner", suggesting that mate choice is playing a role in driving constraint among genes depleted for PTVs.
The main man himself, Charles Darwin, wrote about the relationship between sexual selection and evolution way back in 1871. So, our idea is not new, but our findings represent strong evidence that Darwin’s theory is shaping the gene pool of contemporary human populations.
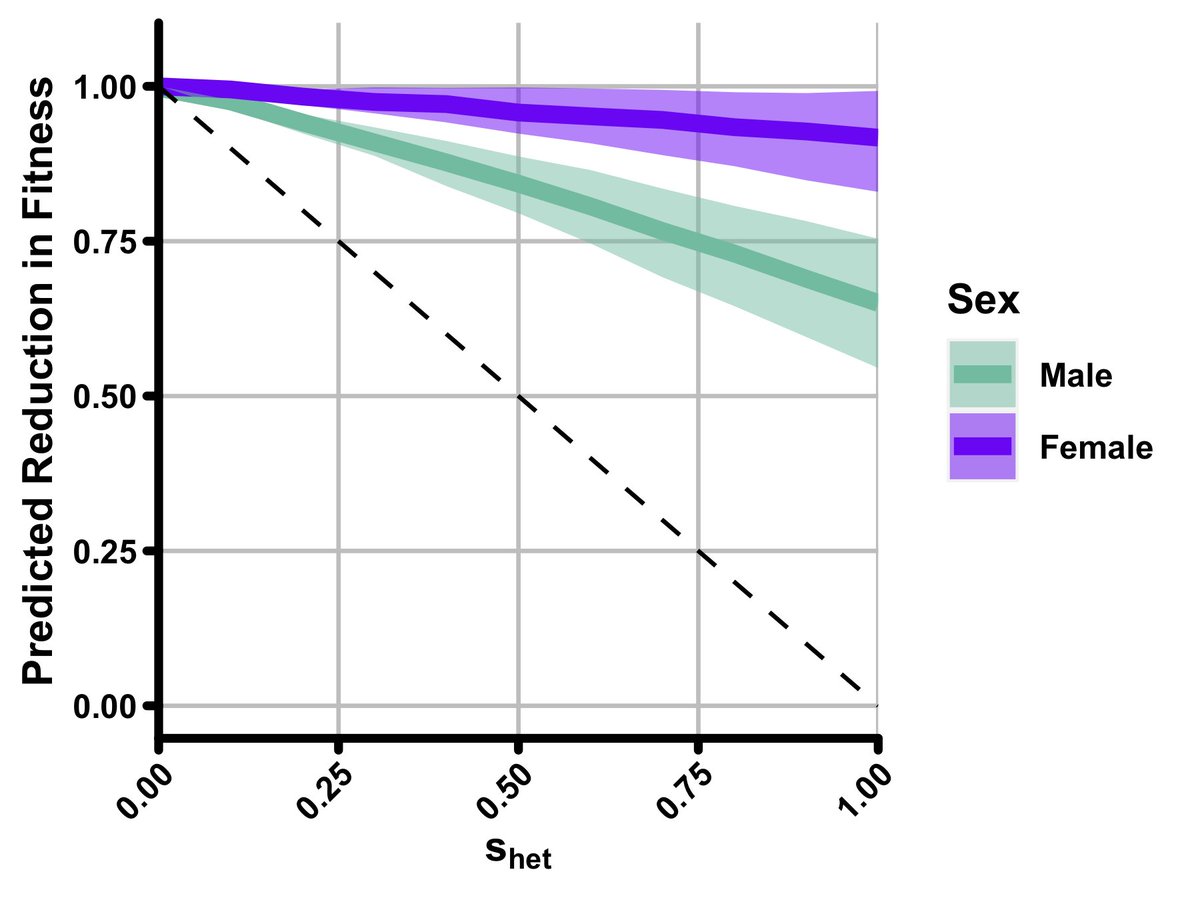
Ultimately, we found that s-het- burden likely explains ~22% of reduced fitness expected by s-het-.
Obviously, there are some biases in the UKBB data and would love to see this in the context of other populations.
Obviously, there are some biases in the UKBB data and would love to see this in the context of other populations.
Finally – a big thanks to our collaborators on the project!
@ma_niemi, @GeorgeKirov1, @BarclayKieron, and @MartinKolk
@ma_niemi, @GeorgeKirov1, @BarclayKieron, and @MartinKolk

 Read on Twitter
Read on Twitter