For the past 4 years I& #39;ve mostly worked on disaggregation regression. As this work is mostly published or in preprint, and because I doubt I& #39;ll be working much more on it, I thought I& #39;d do a thread covering the work.
Disaggregation regression is regression where the response data is at course resolution and the covariates or random effects are at a higher resolution.
Here resolution typically refers to space, but it could be time, phylogeny, or anything else. All of my work has been spatial. I& #39;d love to see someone apply disaggregation regression to phylogeny though!
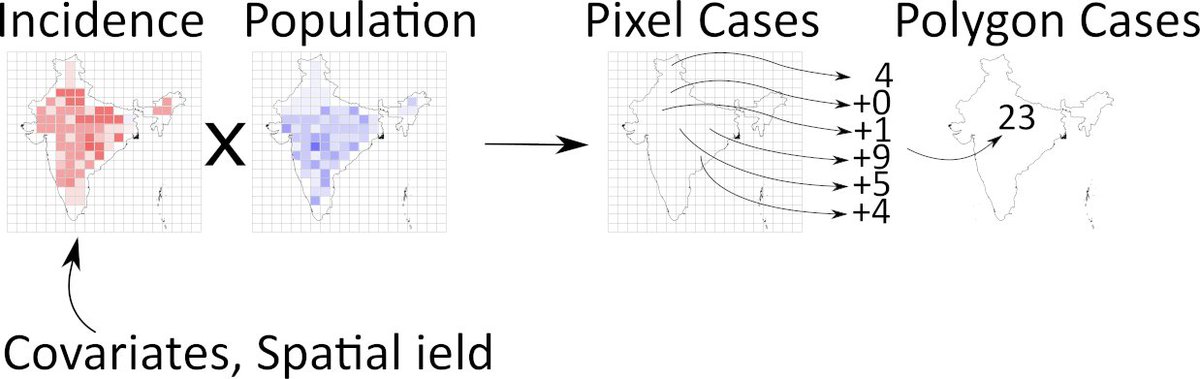
The basic model, with high res variables referenced with j and low res bars with I, looks like this:
yj ~ Pois(incj)
incj = sum( inci x popi )
inci = exp(bXi + spatial_fieldi).
The summation is over all pixels i, in low res area j.
yj ~ Pois(incj)
incj = sum( inci x popi )
inci = exp(bXi + spatial_fieldi).
The summation is over all pixels i, in low res area j.
Generally, standard software can& #39;t fit these types of models. We have been using TMB and @aknandi had written an R package to make the models more accessible.
https://github.com/aknandi/disaggregation
https://github.com/aknandi/d... href=" https://arxiv.org/abs/2001.04847 ">https://arxiv.org/abs/2001....
https://github.com/aknandi/disaggregation
If you want a linear link function, and low resolution covariates, you can fit these models in INLA. @Paula_Moraga_ https://www.sciencedirect.com/science/article/pii/S2211675317301318?casa_token=WQ-_QbEl-HYAAAAA:QcS6frINyNsoEDzbcJA5TtQsQSfU47thRXvjcb2F7QnrlxgmT1rcWG_8baVgU2TA1KTnzT8GRL0">https://www.sciencedirect.com/science/a...
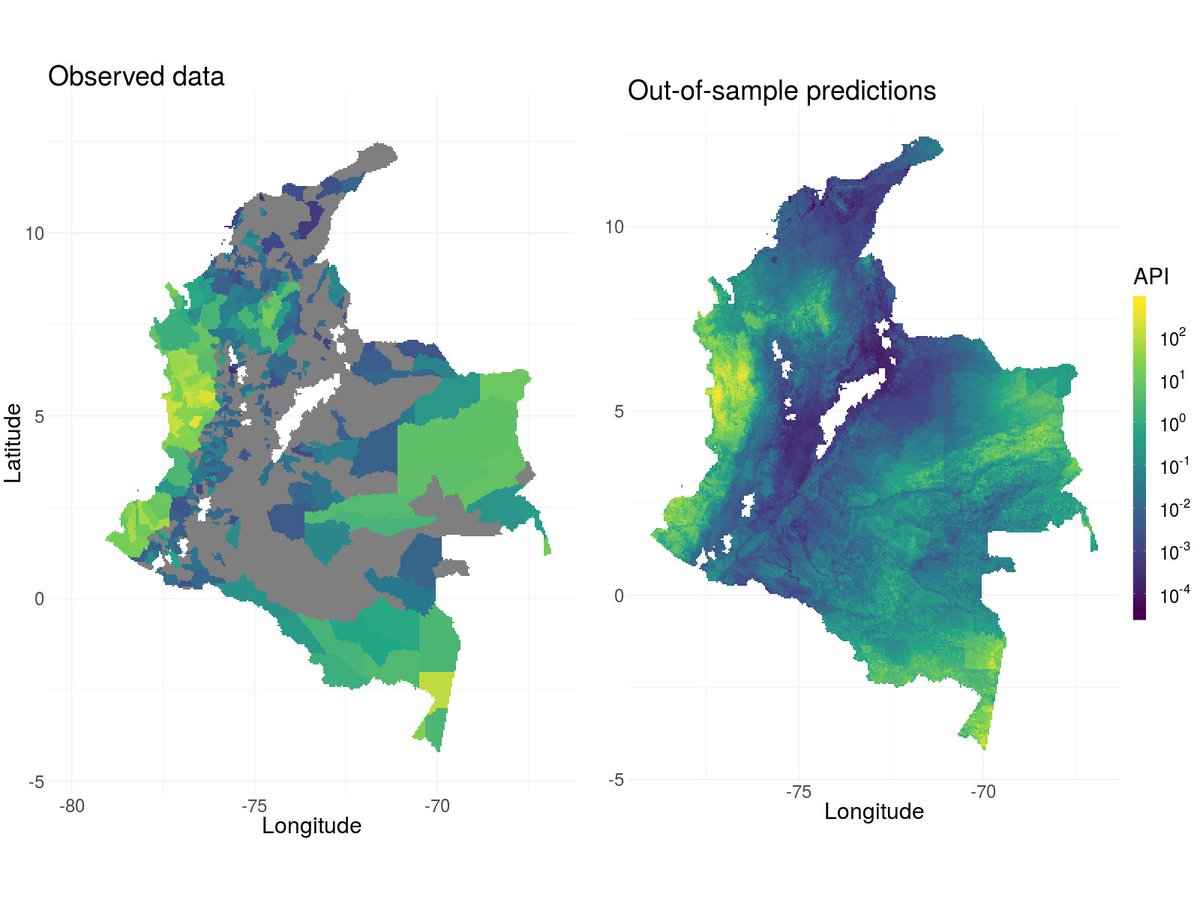
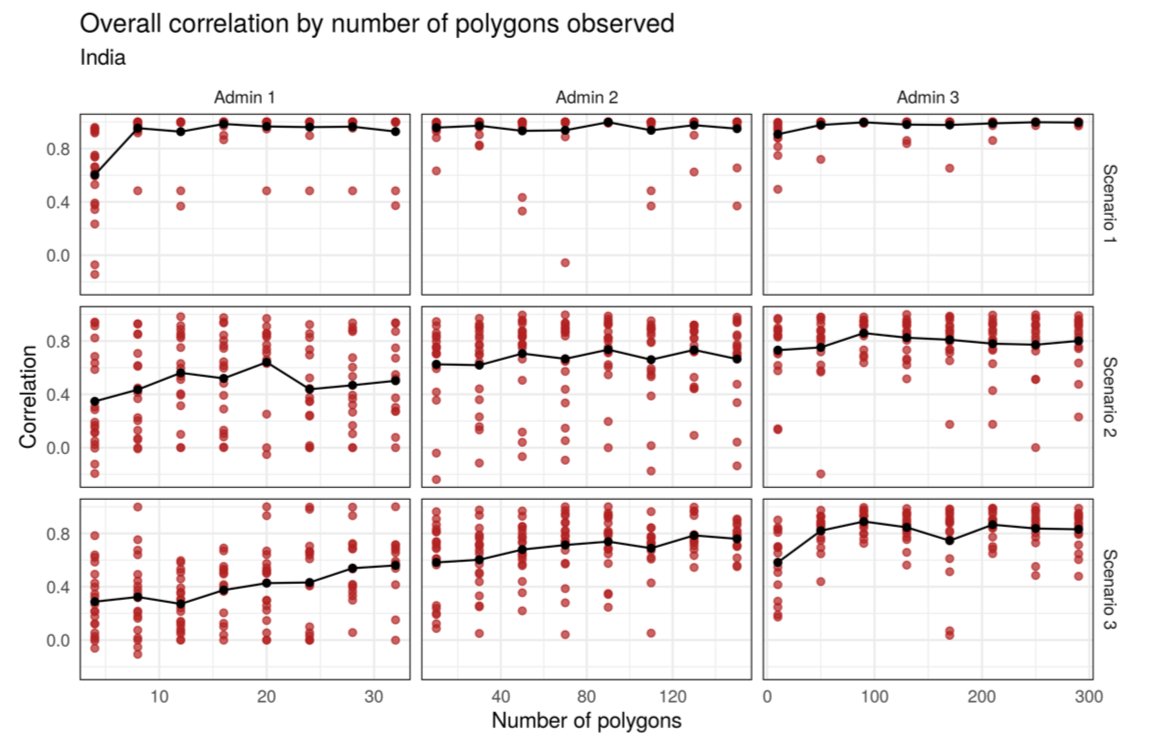
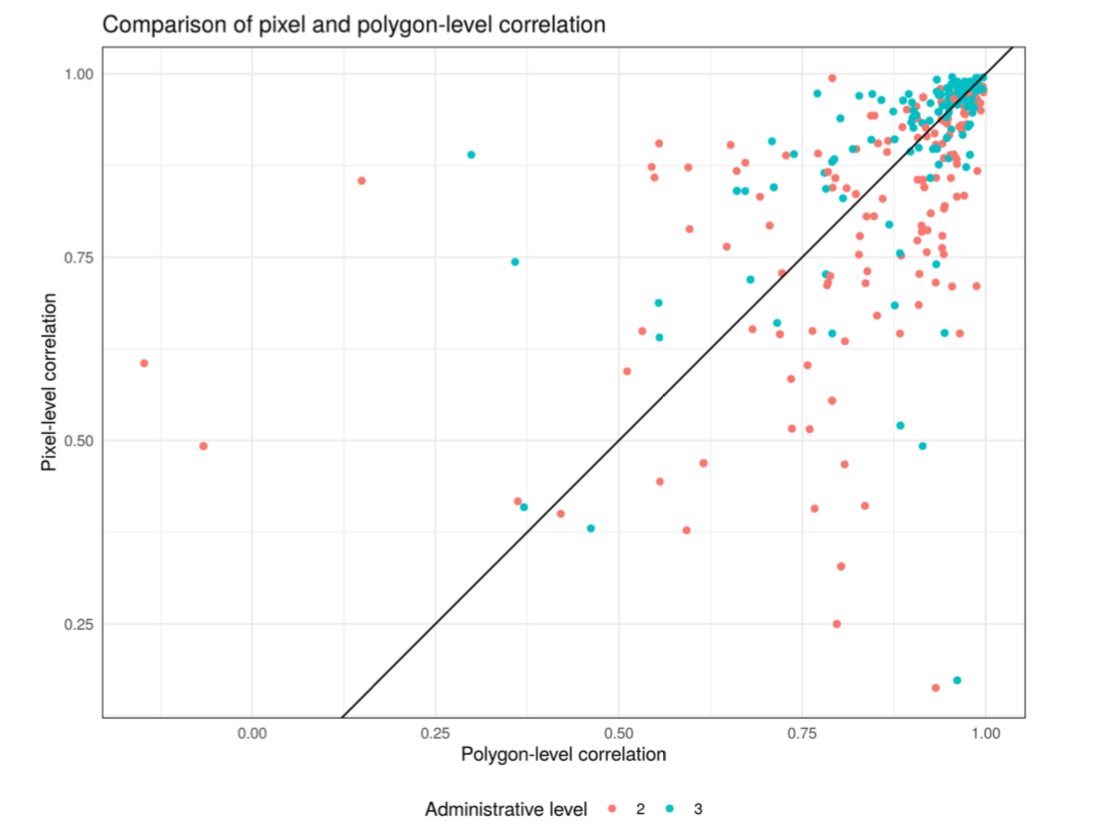
@RohanArambepola did a big simulation study to explore under what circumstances the models work well. He also found low res CV is an ok predictor of high res performance.
https://arxiv.org/abs/2005.03604 ">https://arxiv.org/abs/2005....
https://arxiv.org/abs/2005.03604 ">https://arxiv.org/abs/2005....
We& #39;ve looked at a number of ways to combine low res incidence data with point level prevalence data. We fitted #MachineLearning models to prevalence data and used disaggregation regression on incidence to ensemble the predictions. Modest benefits. https://www.biorxiv.org/content/10.1101/548719v1.abstract">https://www.biorxiv.org/content/1...
We also looked at full joint models of prevalence and incidence data on different spatial scales. This was definitely more finickety. There are benefits (more statistical power, spatial information) and disbenefits (the model learn biases in the prevalence data.
This preprint might be about to undergo quite large changes so take that into account. https://www.medrxiv.org/content/10.1101/2020.02.14.20023069v1">https://www.medrxiv.org/content/1...
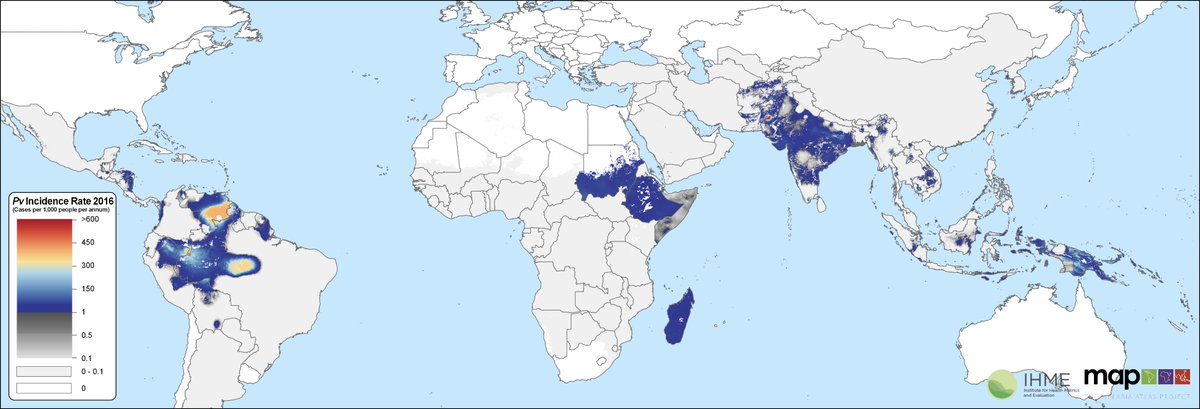
We have applied these models at scale. Here we apply then to predict malaria Vivax globally 2000-2019.
https://www.sciencedirect.com/science/article/pii/S0140673619310967">https://www.sciencedirect.com/science/a...
https://www.sciencedirect.com/science/article/pii/S0140673619310967">https://www.sciencedirect.com/science/a...
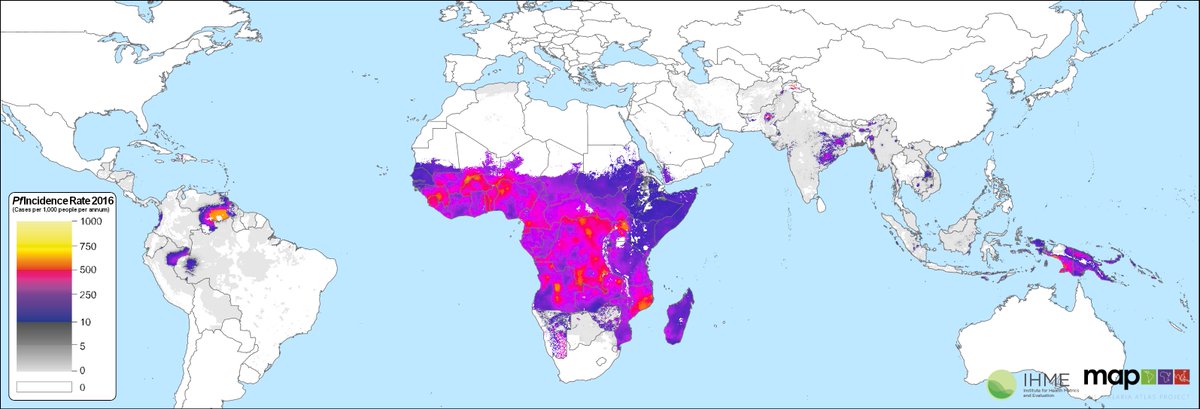
Here we predict malaria falciparum incidence outside of Africa using disaggregation regression. In Africa @DrSamirBhatt primarily used prevalence data.
https://www.sciencedirect.com/science/article/pii/S0140673619310979">https://www.sciencedirect.com/science/a...
https://www.sciencedirect.com/science/article/pii/S0140673619310979">https://www.sciencedirect.com/science/a...
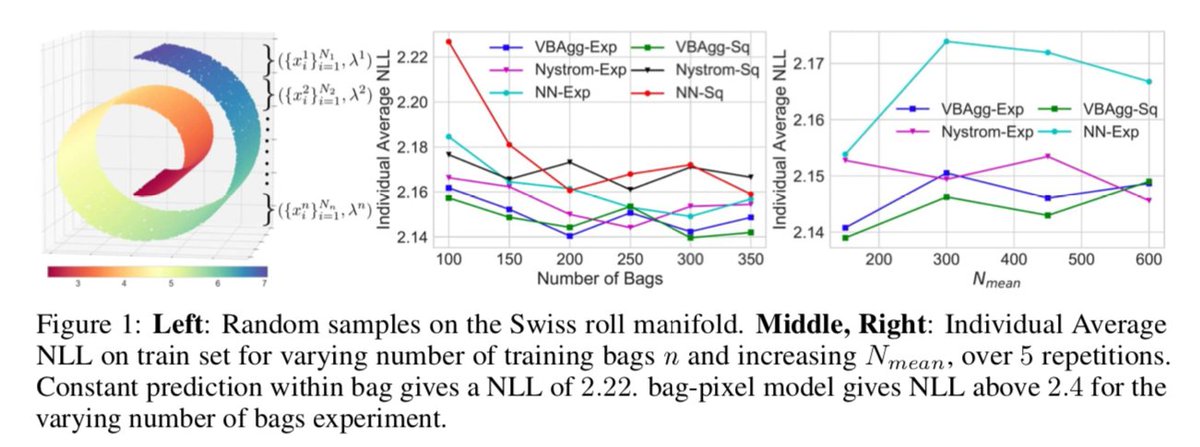
Finally, Leon Law used similar models but using variational Bayes to have a full Gaussian process on covariates and space. The maths is fairly beyond me... I just helped interpret the malaria case study.
http://papers.nips.cc/paper/7847-variational-learning-on-aggregate-outputs-with-gaussian-processes">https://papers.nips.cc/paper/784...
http://papers.nips.cc/paper/7847-variational-learning-on-aggregate-outputs-with-gaussian-processes">https://papers.nips.cc/paper/784...

 Read on Twitter
Read on Twitter