1/n Okay, here& #39;s a long breakdown of the lab& #39;s recent preprint. I have to say I am so excited by this project! SO MUCH WORK by such a great group of trainees (and Laura)!
Basis of Specificity for a Conserved and Promiscuous Chromatin Remodeling Protein
https://www.biorxiv.org/content/10.1101/2020.05.25.115584v1">https://www.biorxiv.org/content/1...
Basis of Specificity for a Conserved and Promiscuous Chromatin Remodeling Protein
https://www.biorxiv.org/content/10.1101/2020.05.25.115584v1">https://www.biorxiv.org/content/1...
This work is the story of how a protein that we& #39;ve been treating as a nonspecific nucleosome spacing and sliding factor is converted into an obligate targeted protein in cells. This mystery has haunted me for years! How does a nonspecific protein become targeted?
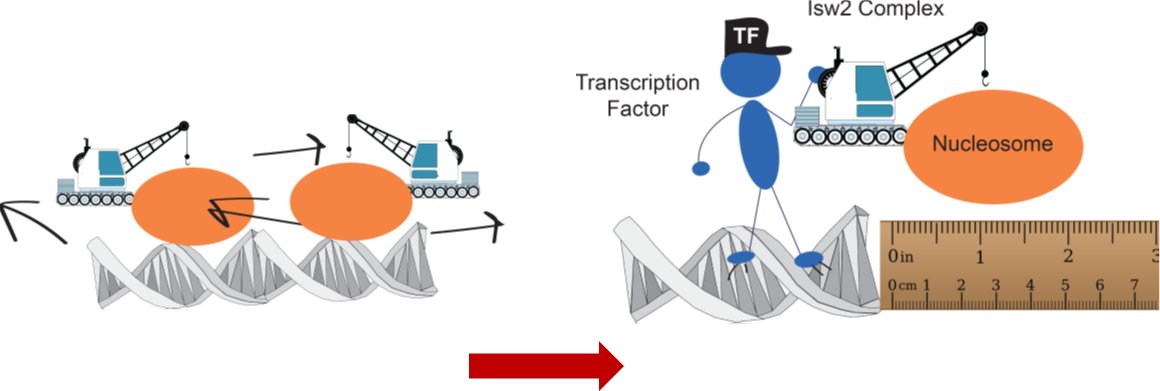
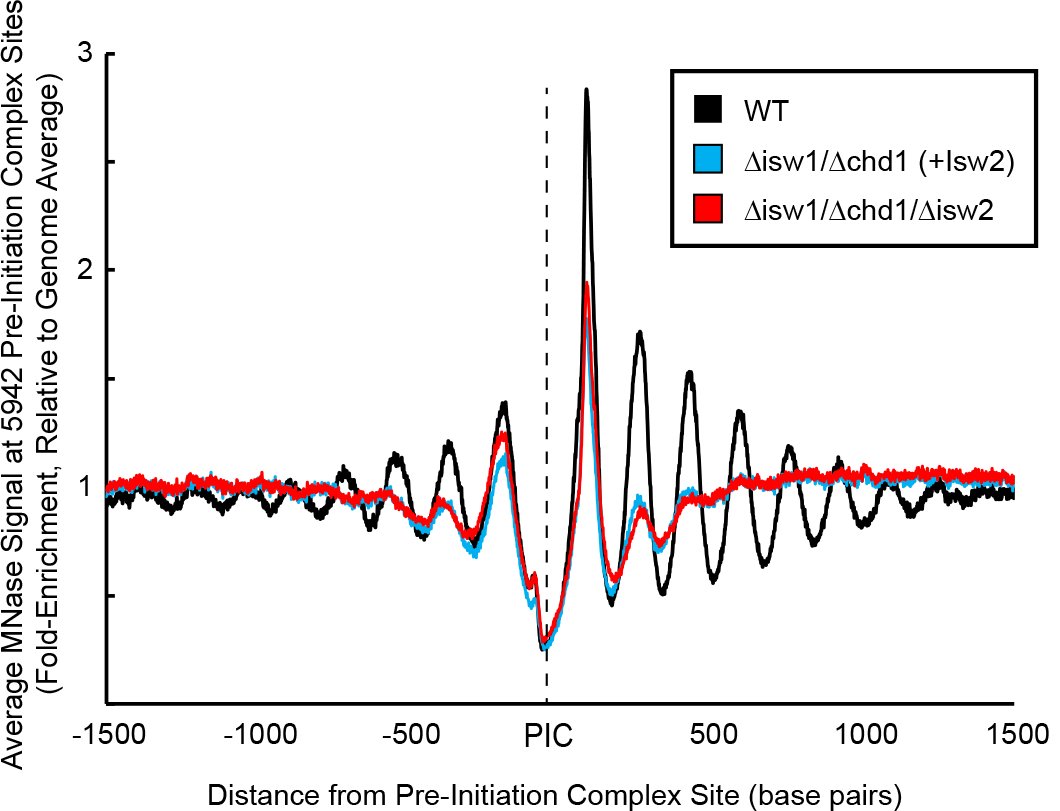
We focused on the conserved Isw2 complex (ACF complex in flies and humans). We wanted to see what our nonspecific protein does in yeast cells. It turns out that there is no global spacing activity (previously seen by the Owen-Hughes and Clark labs).
Instead we see what appear to be very precise and potentially single-nucleosome shifts at sites where Isw2 is stably associated. The first nucleosome gets very well-positioned by Isw2 but subsequent nucleosome structure decays rapidly. Maybe Isw2 only moves single nucleosomes!
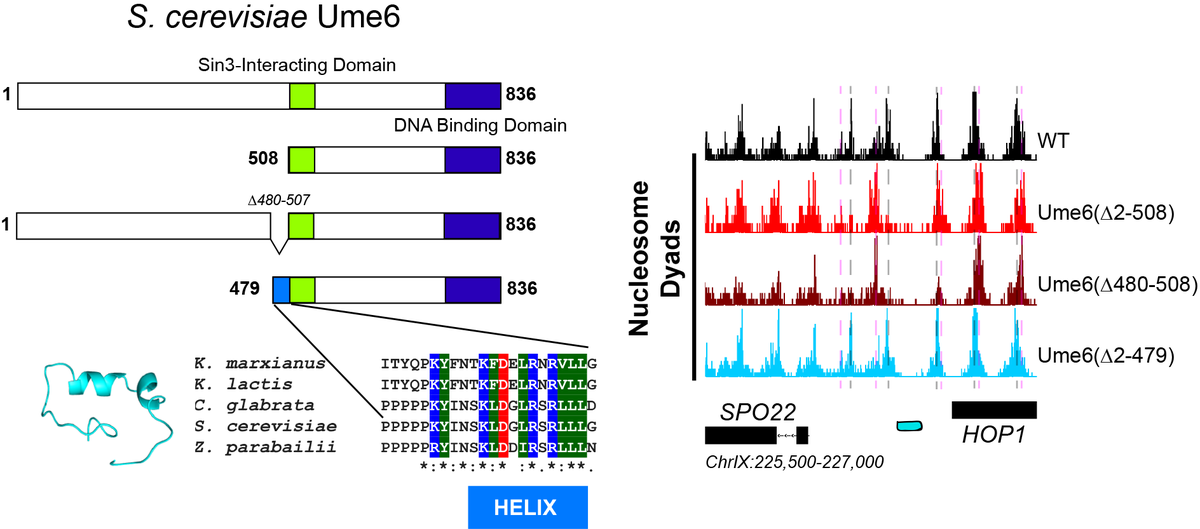
What constrains Isw2 to these single targets? Luckily there is a ton of great work from @ToshioTsukiyama and others showing that the Ume6 repressor has strong genetic interactions with Isw2, and likely interacts physically in cells. Is there an Isw2-recruitment epitope on Ume6?
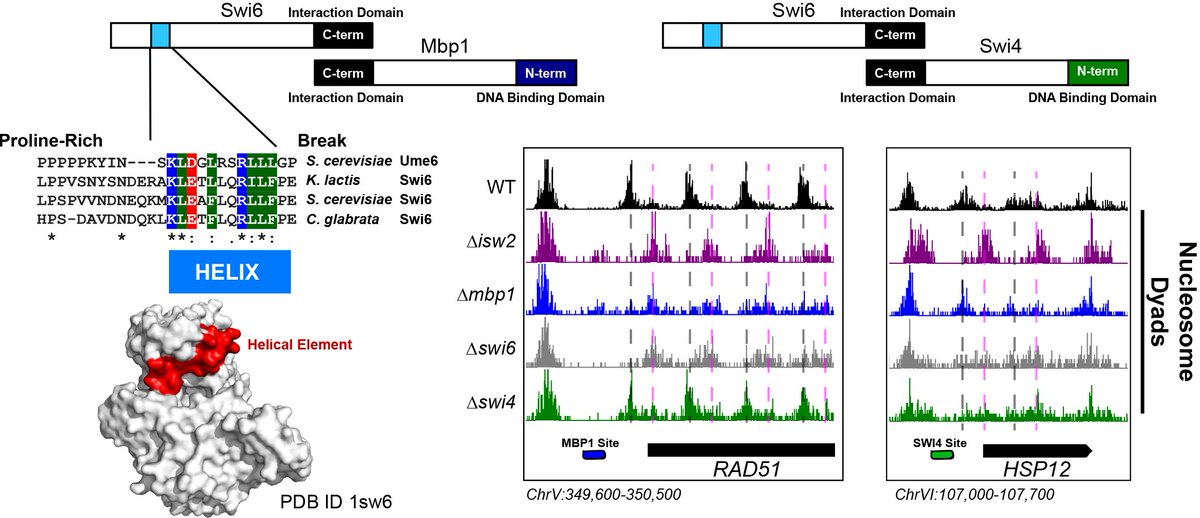
To make a long, painful story short, YES! We narrowed the region down to a conserved portion of Ume6 spanning residues 479-508! When the helix is present, Isw2-directed positions are achieved. When absent, Ume6 binds fine but nucleosomes remain in unshifted (no Isw2) positions.
What& #39;s amazing about this is that other nucleosome spacing factors completely ignore the Ume6 barrier and only Isw2 can move this proximal nucleosome. Also amazing is that this small epitope in Ume6 is enough to organize phased nucleosome arrays through Isw2 and spacing factors!
We were excited to show that this helical element in Ume6 can be displayed ectopically (using SpyCatcher-SpyTag pairs, [thanks Howarth Lab!]). Creating a de novo fusion of the Ume6 helix to a construct that does not recruit Isw2 results in wild-type nucleosome positions!
While working on this project, we found that the cell cycle regulator Mbp1 appears to work with Isw2 at genomic targets. The Isw2 recruitment region was in the Mbp1-interacting protein Swi6, part of the conserved MBF complex. Swi6 has a helix that resembles the Ume6 helix!
Swi6 also forms a complex with Swi4 to create the conserved cell cycle regulatory complex SBF. It turns out that Swi6 also recruits Isw2 activity to Swi4 motifs in the genome as well. So Swi6 is a new adapter protein that brings Isw2 to a subset of Mbp1 and Swi4 sites!
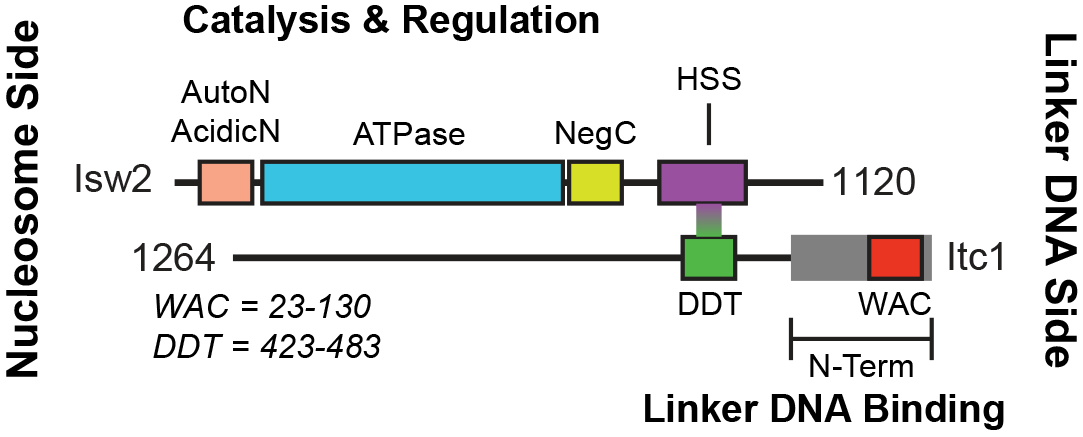
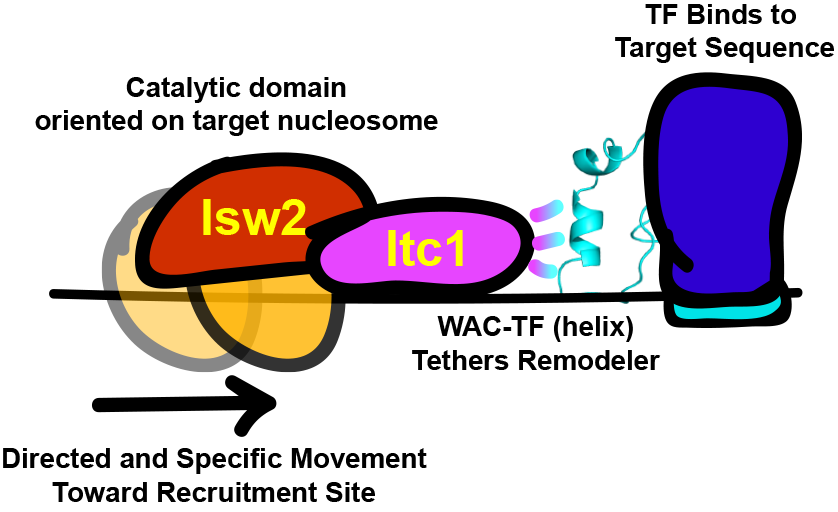
But what part of the Isw2 complex interacts with epitopes in DNA-bound factors? Inspired by work from Frank Pugh& #39;s & Xaiowei Zhuang& #39;s groups, we expected the noncatalytic subunit Itc1 (Acf1 in flies, humans) to be in the right position on chromatin for a targeting function.
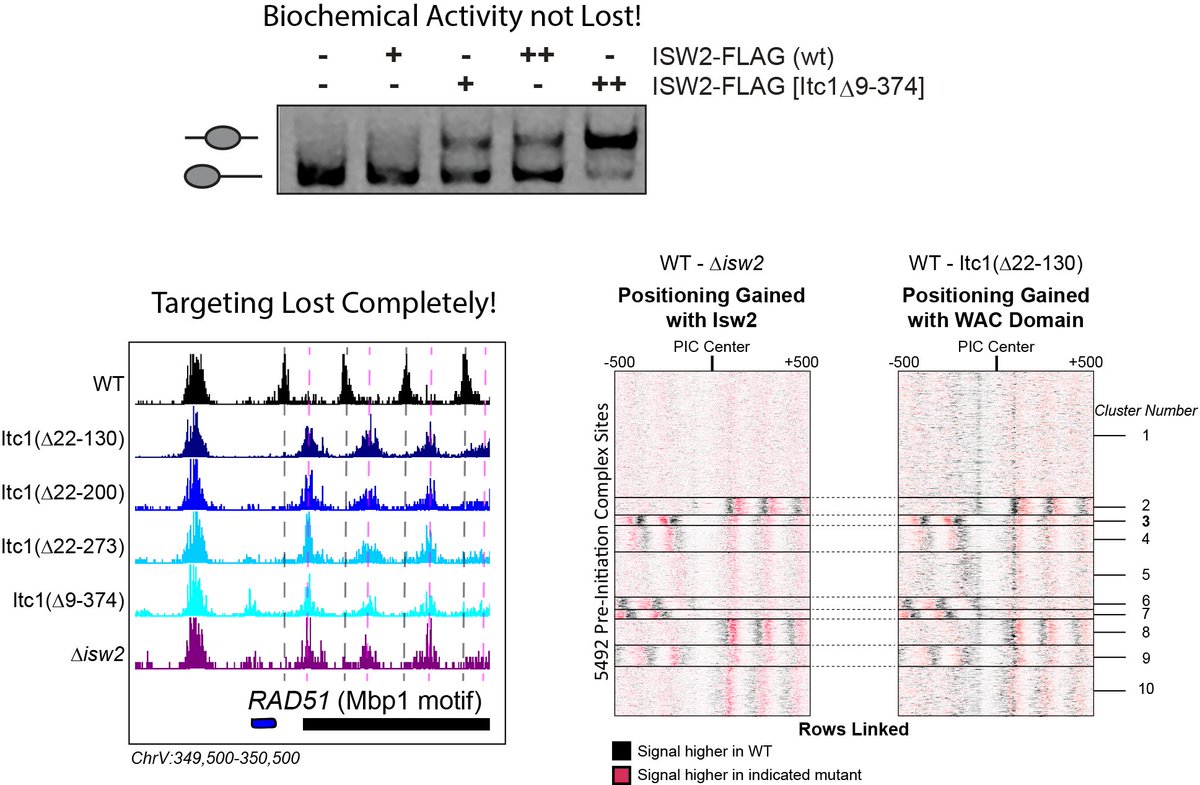
The Zhuang lab found deletion of the N-terminus of Itc1 is lethal. We deleted this domain in a W303 strain saw no loss in biochemical activity but complete loss of genomic targeting (no lethality). We dissected Itc1 to show that targeting is achieved by the conserved WAC domain.
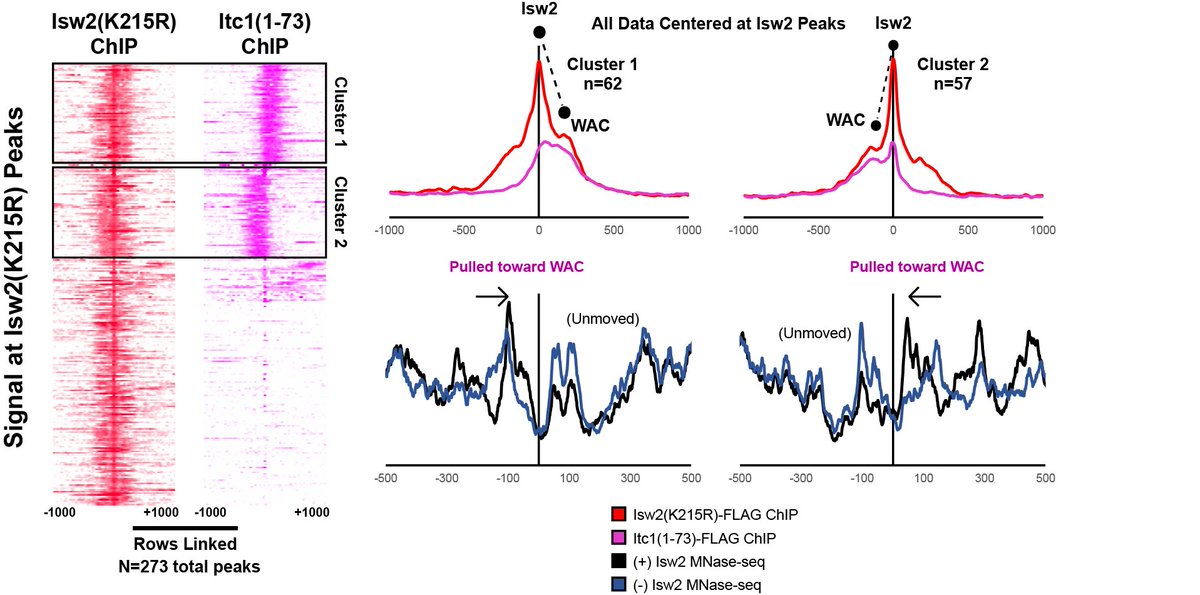
What& #39;s more, a fraction of the conserved WAC domain can bind to Isw2 sites (measured by FLAG ChIP-seq). The Itc1 fragment actually binds distal to the Isw2-selected nucleosome, while the Isw2 catalytic subunit binds on top of the remodeled nucleosome.
This offset nature of Itc1 and Isw2 subunits is consistent with Itc1 being targeted to an epitope or DNA-bound factor to fix the Isw2 catalytic subunit on the proper nucleosome resulting in a pulling motion (tip of hat to David Shore& #39;s group) toward the sequestered Itc1.
Now the sticky part: the ACF complex in Drosophila is clearly a nucleosome spacing factor in cells (shown most recently and clearly by Peter Becker& #39;s group) and the WAC domain is supposed to bind linker DNA, not epitopes in DNA-associated proteins. Why the disconnect?
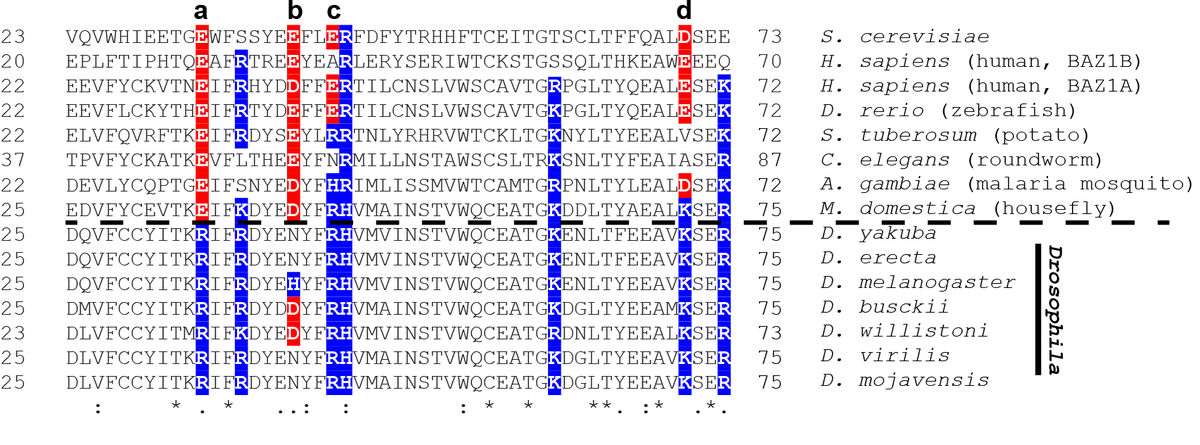
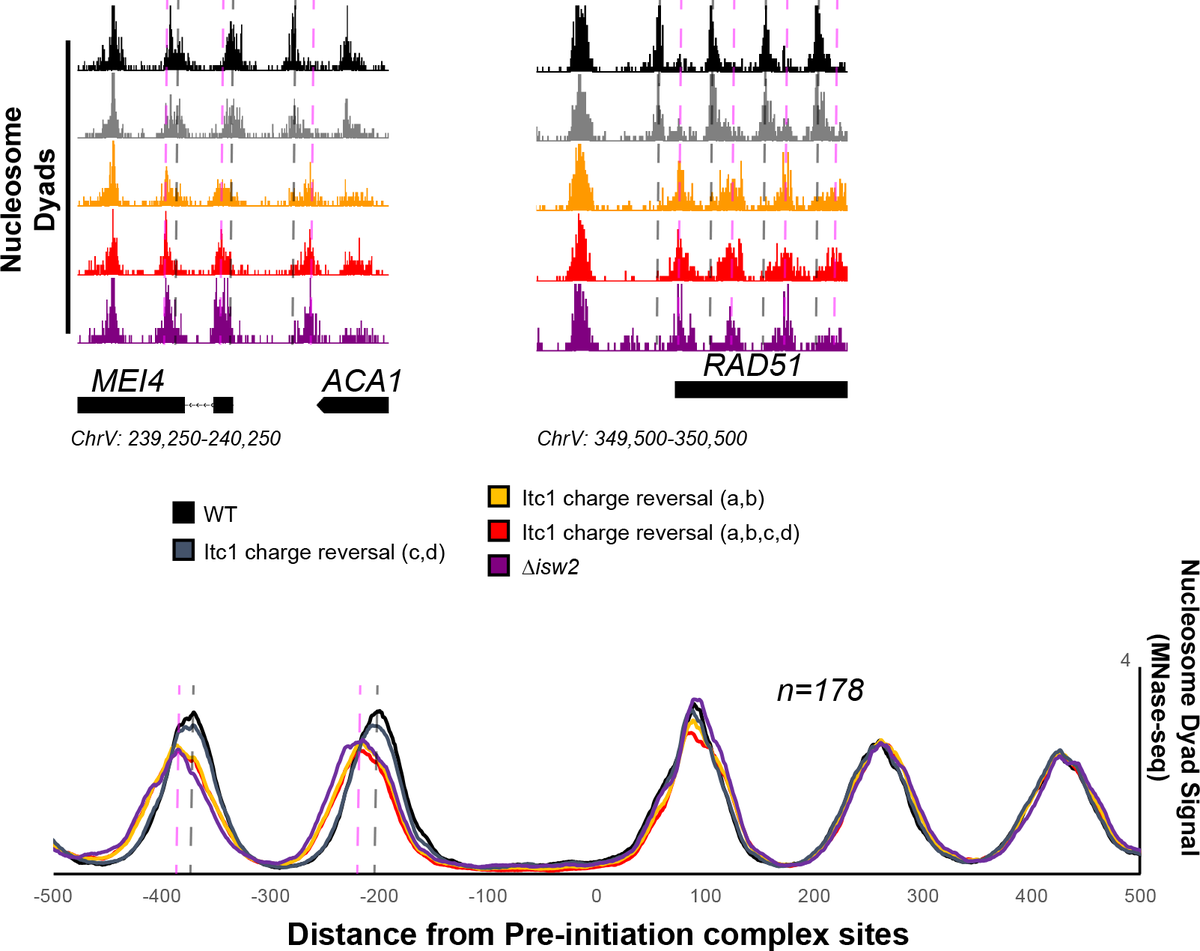
Alignment of the region of the Itc1 WAC domain that binds Isw2 targets highlights that all eukaryotes (I haven& #39;t yet found other counterexamples) possess acidic residues in key locations (here labeled "a" and "b") while the Drosophila genus has several charge reversals (a,b,c,d).
We asked if mutating the Itc1 acidic residues at "a" and "b" was sufficient to abrogate targeting in yeast cells. Sure enough, charge reversal of these two residues is enough to render Isw2 completely inactive at genomic targets!
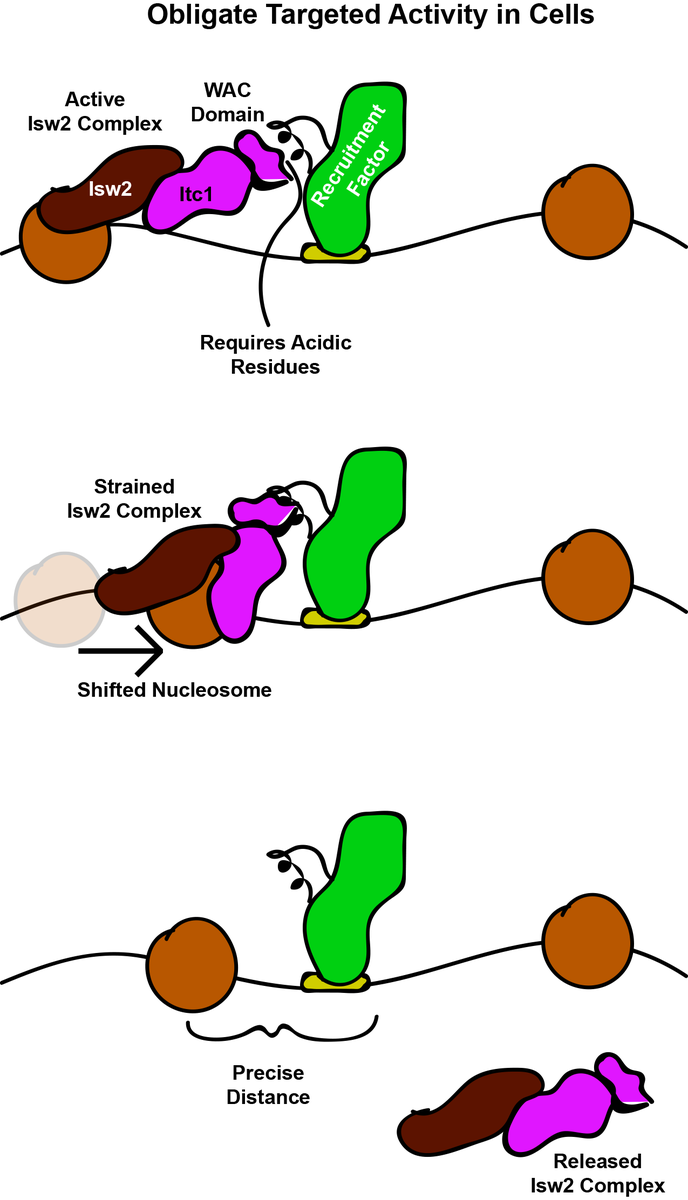
So our model now, is that Itc1 is recruited to DNA-bound factors through small epitopes to constrain Isw2 on target nucleosomes. Isw2 pulls these nucleosomes toward Itc1 until the final precise position is achieved. We speculate what might trigger this release is autoinhibition.
What have we learned? 1) Isw2 (for reasons to be determined in the future) is only active at specific targets in the yeast genome. 2) Targeting is achieved through epitopes on DNA-bound factors interacting with a conserved piece of the Itc1 WAC domain.
3) Swi6 is an Isw2-recruitment adapter that brings Isw2 to a subset of MBF and SBF sites, potentially through a similar epitope found in Ume6. 4) Acidic residues in the WAC domain help target and orient Isw2, but these were lost in Drosophila (and only Drosophila it seems).
Important takeaways and conjectures (at least I think so): 1) The cellular function is not consistent with the biochemical function of Isw2. Perhaps future biochemical experiments will benefit from adding cellular context to tease out why this is so.
2) Small epitopes in DNA-bound factors interact with Itc1 to create an "interacting barrier" that helps Isw2 know which nucleosomes to move and where to stop with respect to the barrier. We think all yeast Isw2 targets will be explained by to-be-found epitope-Itc1 interactions.
3) The WAC domain may have a conserved function in all organisms except Drosophila based on the conservation of key acidic residues. The Owen-Hughes group (and others) have shown that loss of ISWI activity leads to movement of nucleosomes away from factor sites in human cells.
4) We likely missed this function of the WAC domain because strong biochemical and cellular (genomic) work has been performed in Drosophila, the outlier with true nonspecific packing function (possibly because of increased positive charge in the WAC domain).
/end
/end

 Read on Twitter
Read on Twitter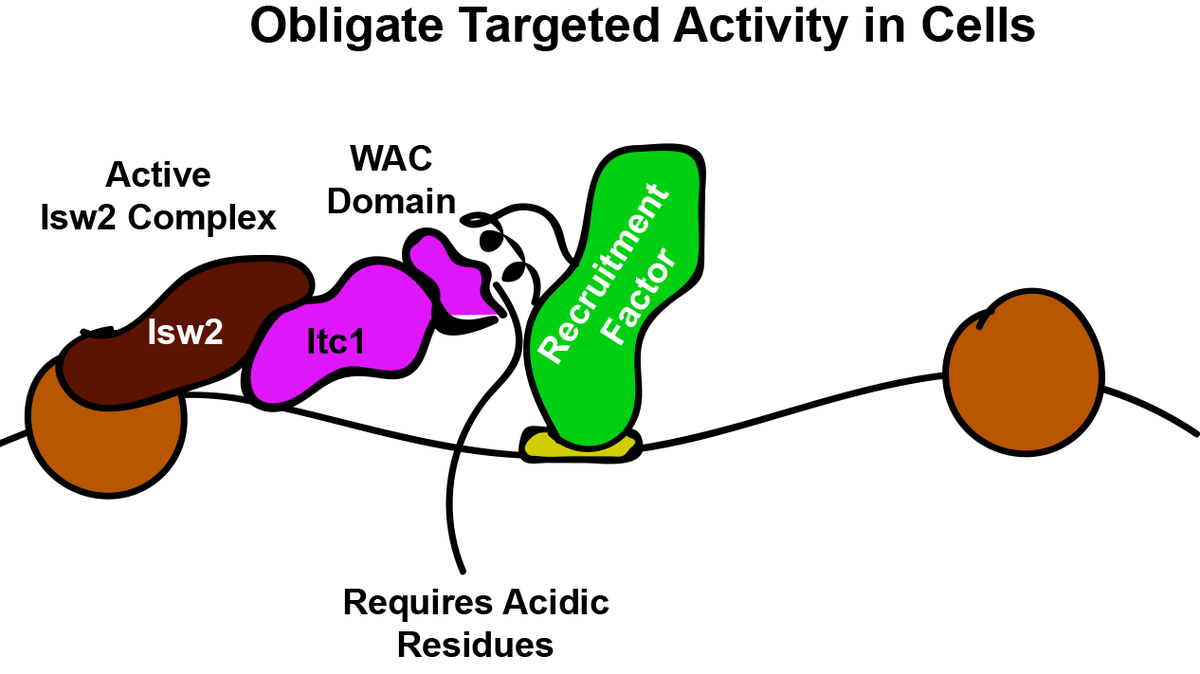

![We were excited to show that this helical element in Ume6 can be displayed ectopically (using SpyCatcher-SpyTag pairs, [thanks Howarth Lab!]). Creating a de novo fusion of the Ume6 helix to a construct that does not recruit Isw2 results in wild-type nucleosome positions! We were excited to show that this helical element in Ume6 can be displayed ectopically (using SpyCatcher-SpyTag pairs, [thanks Howarth Lab!]). Creating a de novo fusion of the Ume6 helix to a construct that does not recruit Isw2 results in wild-type nucleosome positions!](https://pbs.twimg.com/media/EZDF-wWUcAAEeNx.png)