My paper is out on bioRxiv!  https://abs.twimg.com/emoji/v2/... draggable="false" alt="🥳" title="Partying face" aria-label="Emoji: Partying face">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🥳" title="Partying face" aria-label="Emoji: Partying face">
https://www.biorxiv.org/content/10.1101/2020.05.25.114553v1
In">https://www.biorxiv.org/content/1... this work with @dbikard, @epcrocha and others, we used CRISPRi screening in a collection of E. coli isolates to better define essential genes at the species level.
Follow this thread for a summary (1/8)
 https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Down pointing backhand index" aria-label="Emoji: Down pointing backhand index">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Down pointing backhand index" aria-label="Emoji: Down pointing backhand index">
https://www.biorxiv.org/content/10.1101/2020.05.25.114553v1
In">https://www.biorxiv.org/content/1... this work with @dbikard, @epcrocha and others, we used CRISPRi screening in a collection of E. coli isolates to better define essential genes at the species level.
Follow this thread for a summary (1/8)
Most genomics studies are focused on single model strains which are not representative of the microbial diversity of the species. As a result, the mechanisms by which gene essentiality evolves at a small scale remain vague. (2/8)
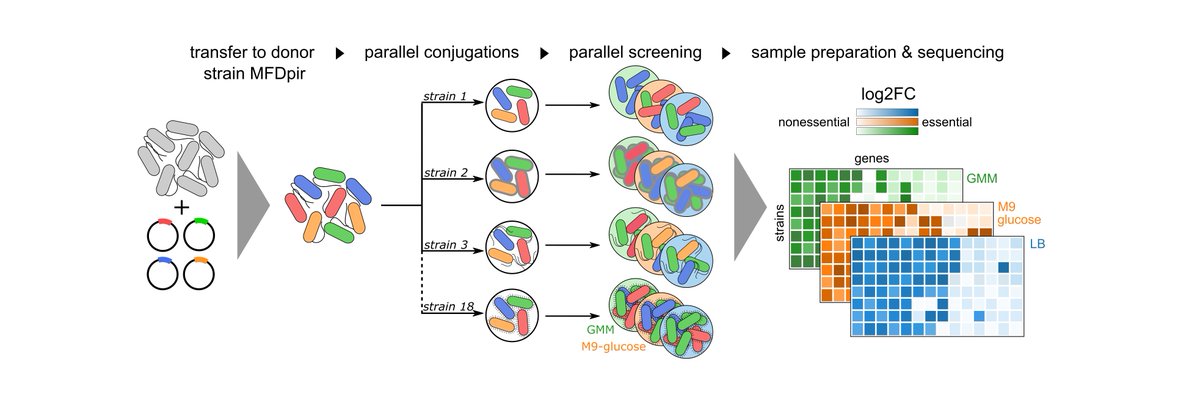
Here, we develop a new CRISPRi vector that works in most E. coli natural isolates as well as in closely-related species. We then designed a sgRNA library to target ~3400 persistent genes of the E. coli species and showed that it can confidently predict essential genes. (3/8)
We used it to screen 18 different isolates from various origins and lifestyles in 3 growth conditions. We found LOTS of differences in essentiality between strains.
This shows that gene essentiality can quickly evolve at the strain level. (4/8)
This shows that gene essentiality can quickly evolve at the strain level. (4/8)
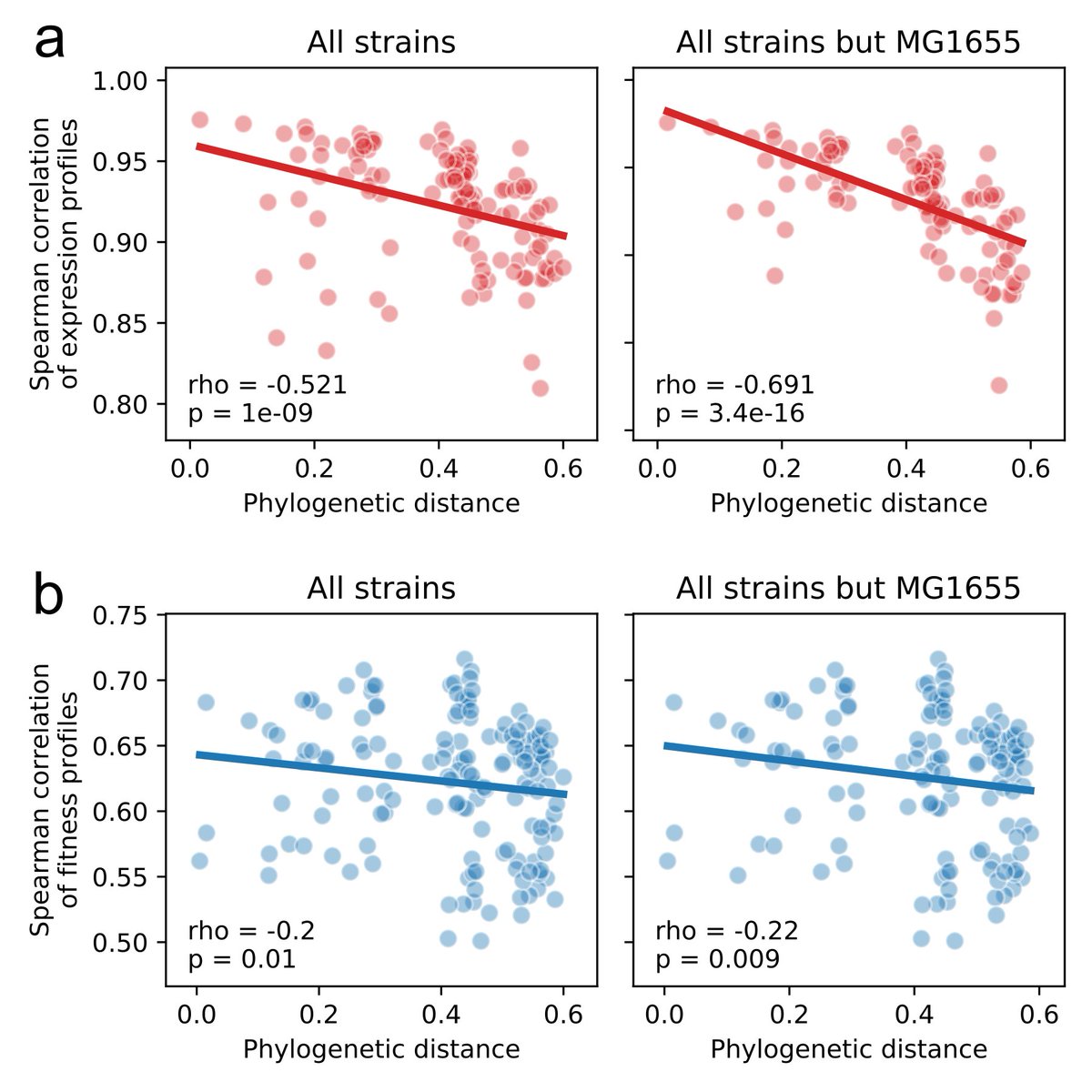
We also performed RNAseq and found that changes in expression level of core genes correlates very well with phylogeny, but changes in essentiality do not. (5/8)
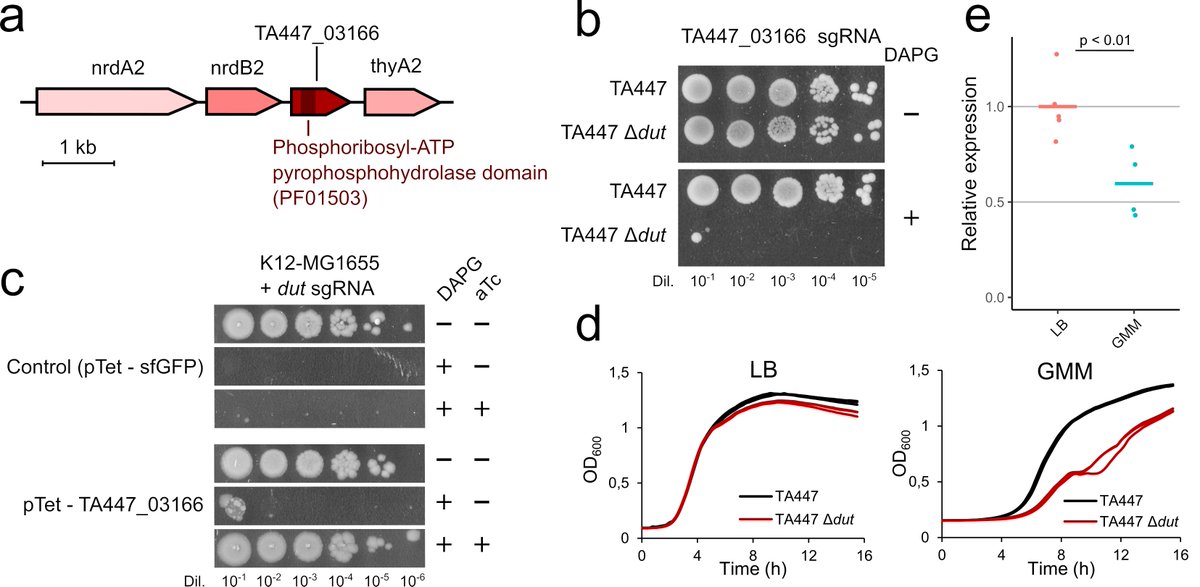
We then studied the mechanisms explaining these variations. We found few cases where the acquisition of homologs or analogs on a mobile genetic element makes a gene nonessential. Here is the example of a dUTPase that makes dut nonessential  https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Down pointing backhand index" aria-label="Emoji: Down pointing backhand index"> (6/8)
https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Down pointing backhand index" aria-label="Emoji: Down pointing backhand index"> (6/8)
Conversely, we found that systems encoded on mobile genetic elements can make persistent genes essential!
This shows that many nonessential genes can become essential on a recurrent basis upon horizontal transfer of some genetic systems. We validated such 2 systems https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Down pointing backhand index" aria-label="Emoji: Down pointing backhand index"> (7/8)
https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Down pointing backhand index" aria-label="Emoji: Down pointing backhand index"> (7/8)
This shows that many nonessential genes can become essential on a recurrent basis upon horizontal transfer of some genetic systems. We validated such 2 systems
Our study shows that genetic diversity influences gene essentiality and that genomics studies should include various strains of the same species. It is also an example of how CRISPRi screens can interrogate genes in high-throughput at a low cost.
Thanks to all authors https://abs.twimg.com/emoji/v2/... draggable="false" alt="🤗" title="Hugging face" aria-label="Emoji: Hugging face"> (8/8)
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🤗" title="Hugging face" aria-label="Emoji: Hugging face"> (8/8)
Thanks to all authors

 Read on Twitter
Read on Twitter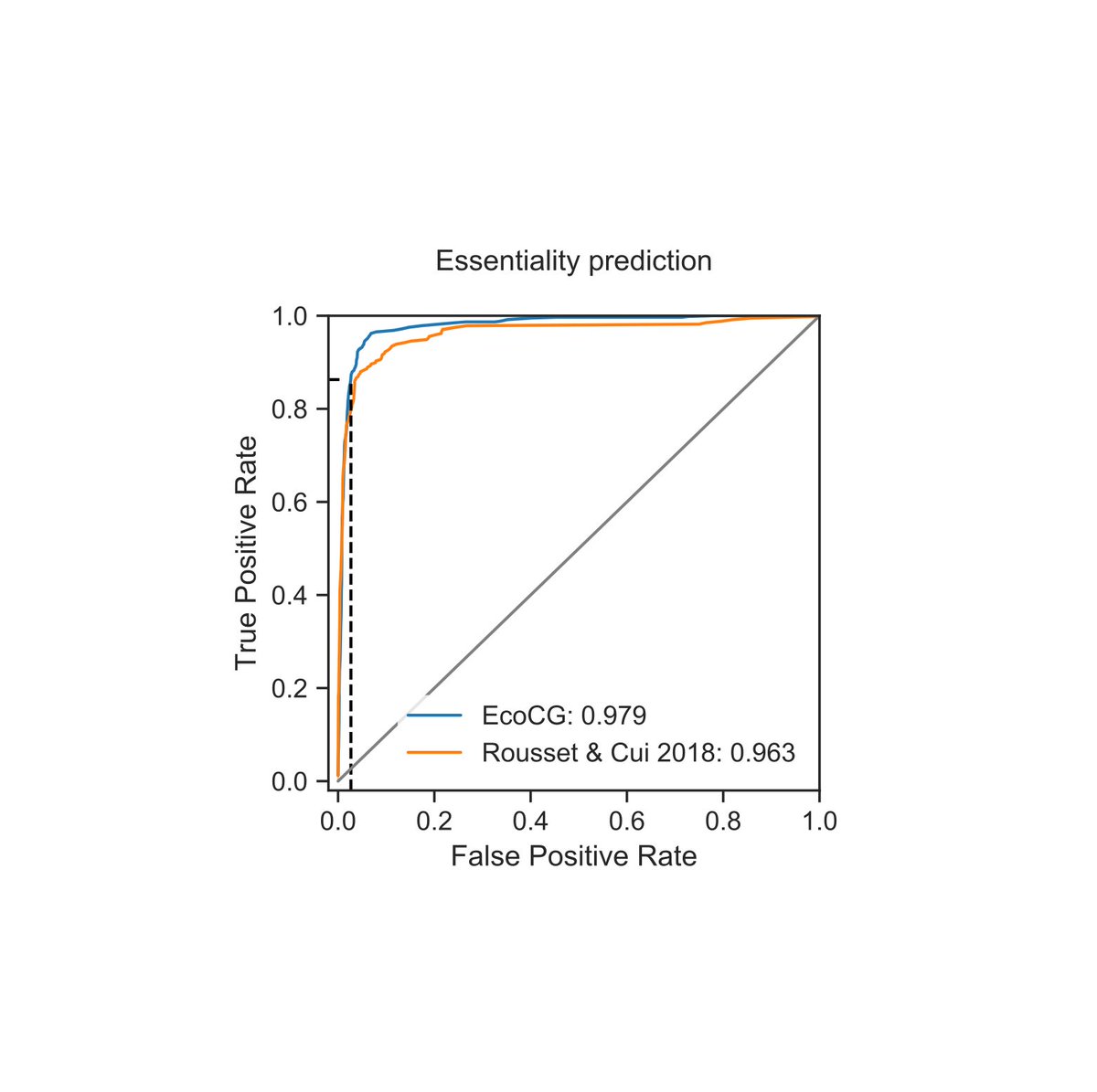
 (6/8)" title="We then studied the mechanisms explaining these variations. We found few cases where the acquisition of homologs or analogs on a mobile genetic element makes a gene nonessential. Here is the example of a dUTPase that makes dut nonessential https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Down pointing backhand index" aria-label="Emoji: Down pointing backhand index"> (6/8)" class="img-responsive" style="max-width:100%;"/>
(6/8)" title="We then studied the mechanisms explaining these variations. We found few cases where the acquisition of homologs or analogs on a mobile genetic element makes a gene nonessential. Here is the example of a dUTPase that makes dut nonessential https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Down pointing backhand index" aria-label="Emoji: Down pointing backhand index"> (6/8)" class="img-responsive" style="max-width:100%;"/>
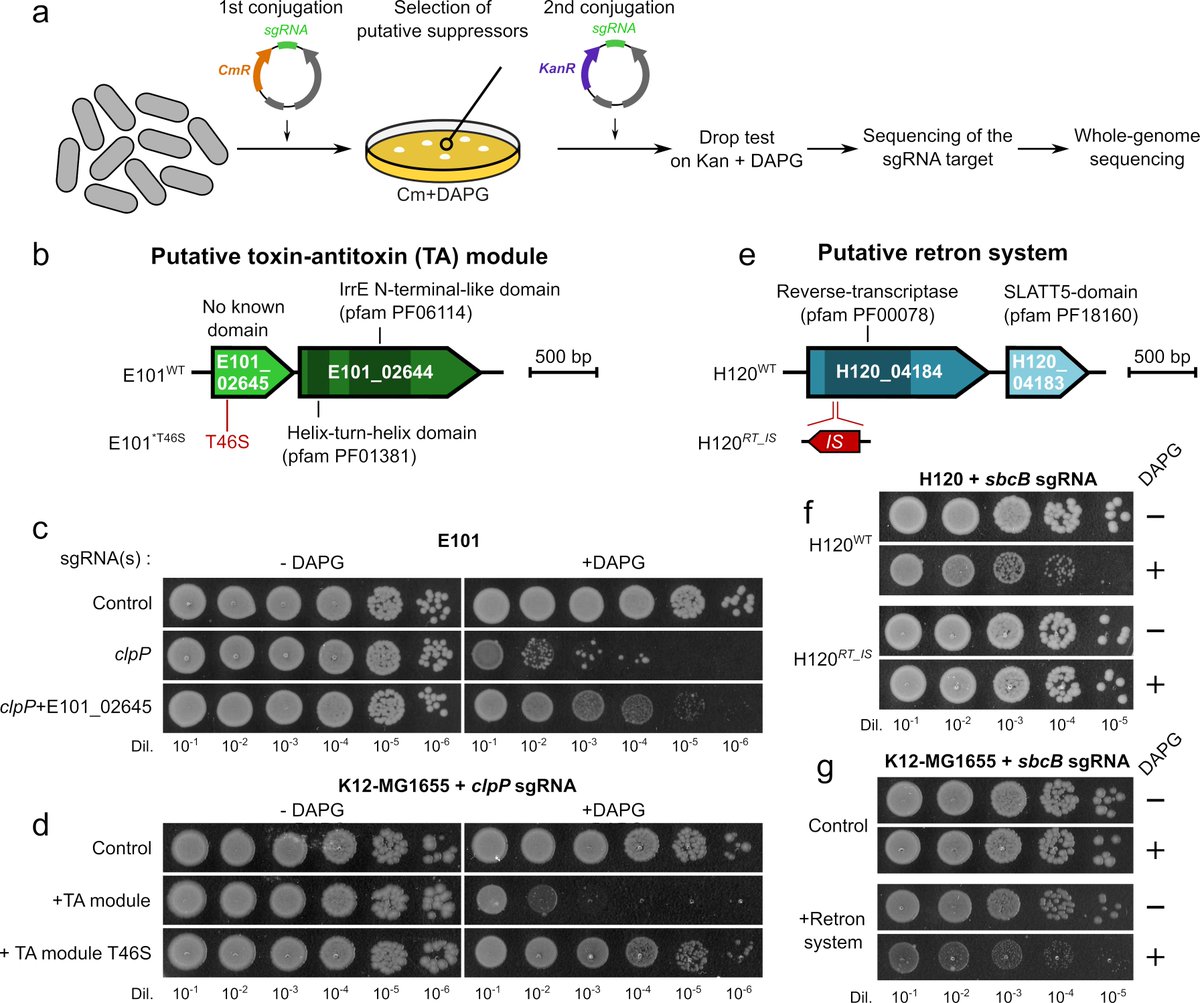 (7/8)" title="Conversely, we found that systems encoded on mobile genetic elements can make persistent genes essential! This shows that many nonessential genes can become essential on a recurrent basis upon horizontal transfer of some genetic systems. We validated such 2 systems https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Down pointing backhand index" aria-label="Emoji: Down pointing backhand index"> (7/8)" class="img-responsive" style="max-width:100%;"/>
(7/8)" title="Conversely, we found that systems encoded on mobile genetic elements can make persistent genes essential! This shows that many nonessential genes can become essential on a recurrent basis upon horizontal transfer of some genetic systems. We validated such 2 systems https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Down pointing backhand index" aria-label="Emoji: Down pointing backhand index"> (7/8)" class="img-responsive" style="max-width:100%;"/>


