Hungry for new high-throughput methods? Take a bite out of SAMOSA! The first preprint from the Ramani Lab, brilliantly led by grad student @NourAbdulhay and PD @cpmcnally in my lab, in close collab. with PD Laura Hsieh & Narlikar Lab! Tweetorial below: https://www.biorxiv.org/content/10.1101/2020.05.20.105379v1">https://www.biorxiv.org/content/1...
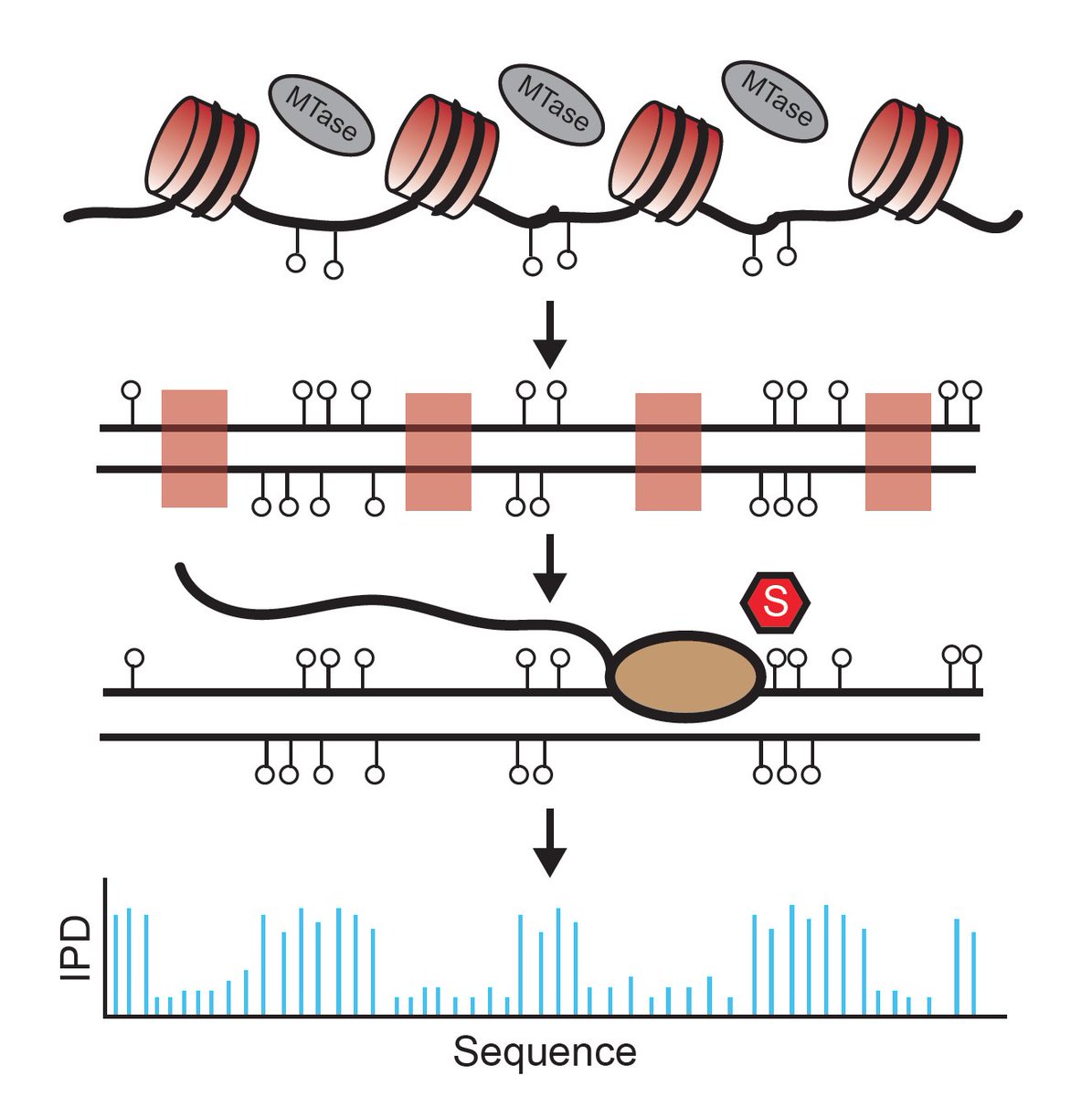
One of our standing goals is to extend massively multiplex measurement to 3rd Gen sequencing tech—this is our first of hopefully a few examples of that. We asked: can we use PacBio SMRT sequencing to footprint oligonucleosomes in a non-destructive way?
We knew from several prior studies that SMRT could natively and robustly detect m6dA via changes in polymerase kinetics, so we wondered if we could selectively adenine-methylate linker DNA (EcoGII) and resolve successive nucleosomes by directly detecting mods on Sequel/Sequel II.
We first tested w/ in vitro assembled Widom 601 chromatin arrays; v. excited by the resolution (not quite base-pair, but close)! After cycling thru unsatisfying assay names, we settled on the Single-molecule Adenine Methylated Oligonucleosome Sequencing Assay—SAMOSA.
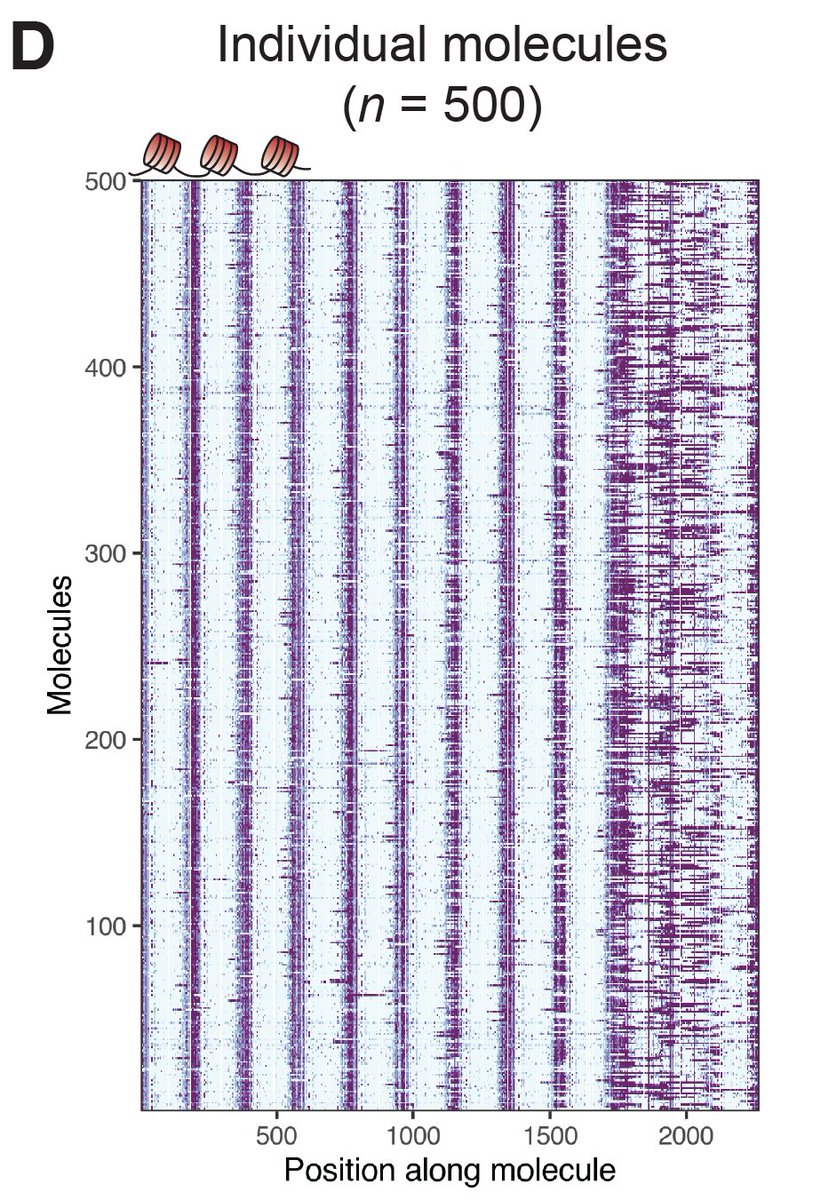
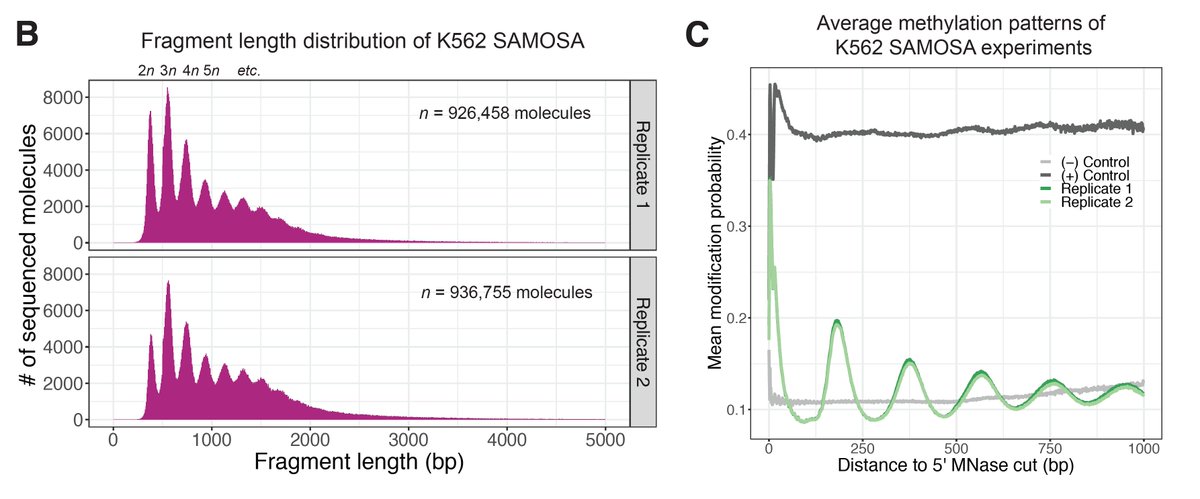
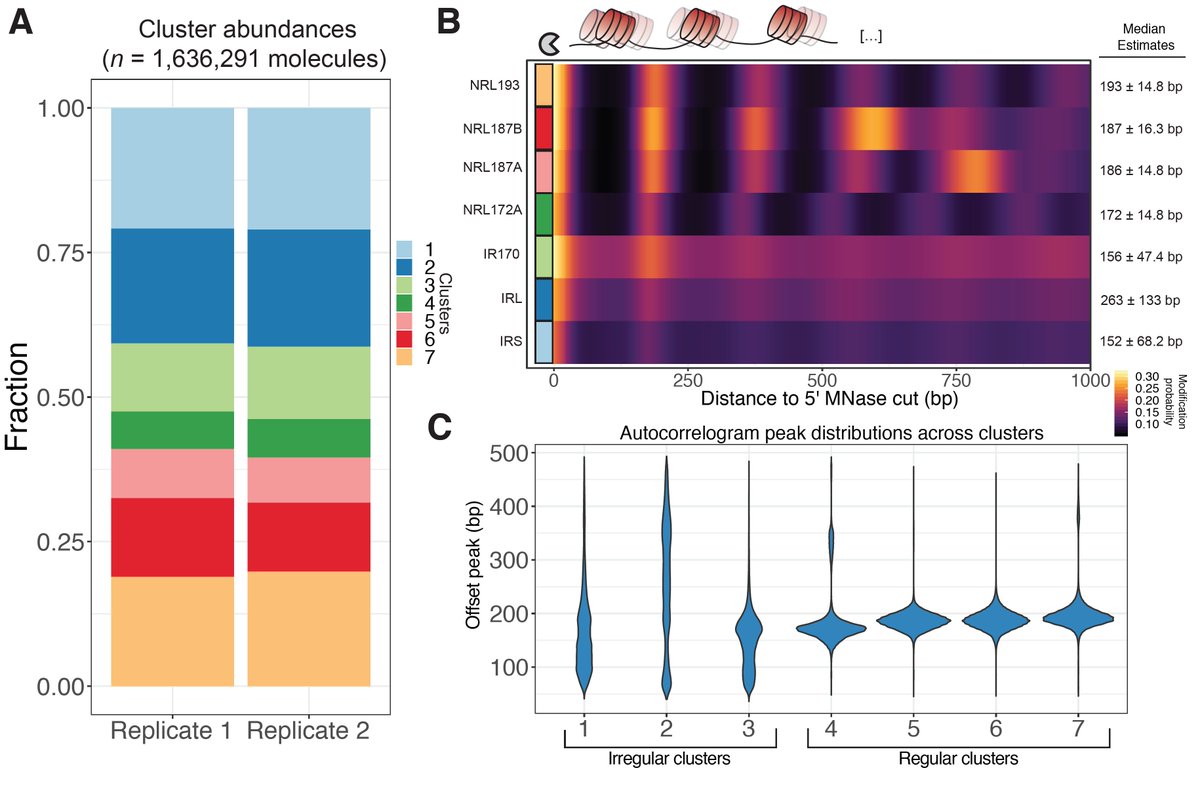
Next applied SAMOSA to arrays from living cells. We borrowed protocol to solubilize MNase-cut oligonucs from mammalian nuclei, & methylated (K562). Approach provides “di-omic” info—MNase cuts @ ends (a la Array-seq of Baldi et al), and m6dA signal to footprint nucs along mol.
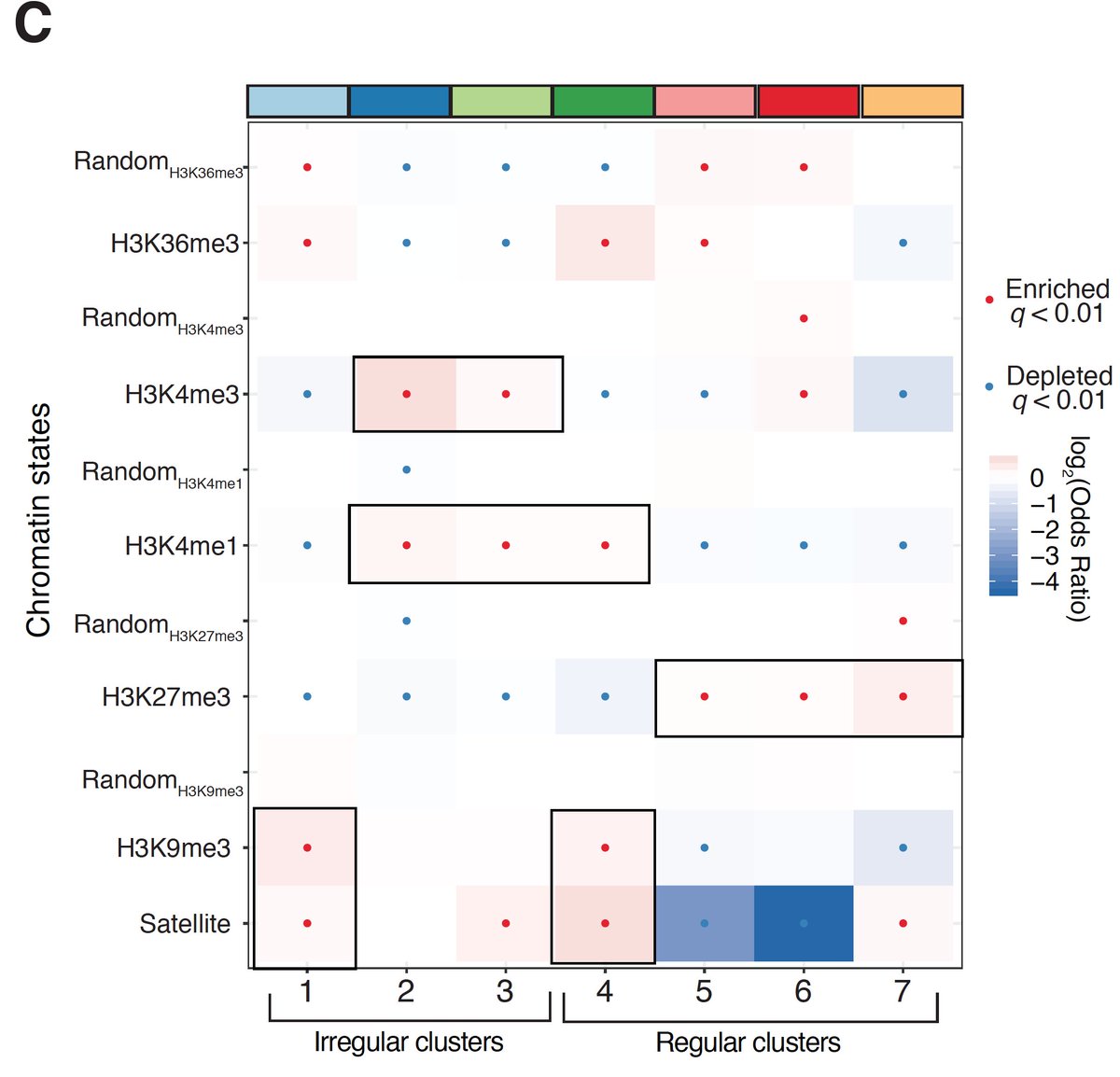
We borrowed an algorithm often used in sc-omics & clustered mols. We found 7 diff. ‘oligonucleosome patterns’ demarcated by both nuc regularity & spacing. We looked @ nuc patterns @ TF binding sites (low coverage but hints of cool bio!), & in epigenomic domains (read preprint!)
A major takeaway from all of our analyses is that chromatin is *very* heterogeneous in terms of pattern usage. Still, we do find that specific chromatin types (e.g. const. heterochromatin, H3K36me3 domains) are punctuated by specific classes of oligonucleosome pattern.
We hope our assay + early analyses provide some valuable context as we all think about how nuc. spacing & regularity give rise to higher-order chromatin org. GEO sub still WIP but *all* processed data is now available—links to data & code can be found @ https://github.com/RamaniLab/SAMOSA">https://github.com/RamaniLab...
gazillion congrats to @NourAbdulhay , who fearlessly spearheaded this project as lab’s first grad student, and who is currently prepping for quals! This work also wouldn’t have happened without Laura & Geeta, as well as the best collabo-neighbor one could ask for in @genophoria.
Plenty of former @UW involved here to thank as well: @JG_Underwood, @SivaKasinathan & co-lead author / former roommate / first lab postdoc @cpmcnally. And @JShendure for constantly telling me that if it works on paper, it’ll eventually work =).
And ofc, this would be impossible w/o the awesome support of the Sandler Fellows Program / @UCSF PBBR / @Ucsf_Biochem!
On a more personal note, excited to see realized an idea that sprung from a ’16 epigenomics conference convo. At the time, heard about @arnaud_kr’s amazing (unpub) dSMF work walking through El Yunque with Dirk Schubeler. Got me thinking about how to avoid GpC / CpG / bisulfite...
m6dA + PacBio seemed like an ‘aha!’ soln.! Wasn’t a great nonspec. m6dA MTase at time, so when I got back to @JShendure lab I purified m6dAse hia5, but proj. got sidelined. Even after EcoGII released, still had throughput issue bc of RSII. Enter Sequel II, and problem solved!
anyway thx for coming to this ted talk. DO also check out some fantastic published papers using the ONT platform to measure long-range chrom. access. in yeast from @zoharshipony / @WJGreenleaf & Kin-Fai Au groups: https://www.nature.com/articles/s41592-019-0730-2">https://www.nature.com/articles/... / https://genome.cshlp.org/content/early/2019/06/14/gr.251116.119,">https://genome.cshlp.org/content/e... as well as ...
... a preprint from @timp0 lab ( https://www.biorxiv.org/content/10.1101/504993v1).">https://www.biorxiv.org/content/1... It’s an exciting time to be developing methods for 3rd Gen sequencing!

 Read on Twitter
Read on Twitter