Finally, our paper on Genomic Profiling of Smoldering Multiple Myeloma is out @JCO @ASCO_pubs! A work that took more than 3 years but with amazing collaborations led to this large study on smoldering multiple myeloma #SMM #mmsm https://ascopubs.org/doi/full/10.1200/JCO.20.00437#.XshAX-ORfn0">https://ascopubs.org/doi/full/...
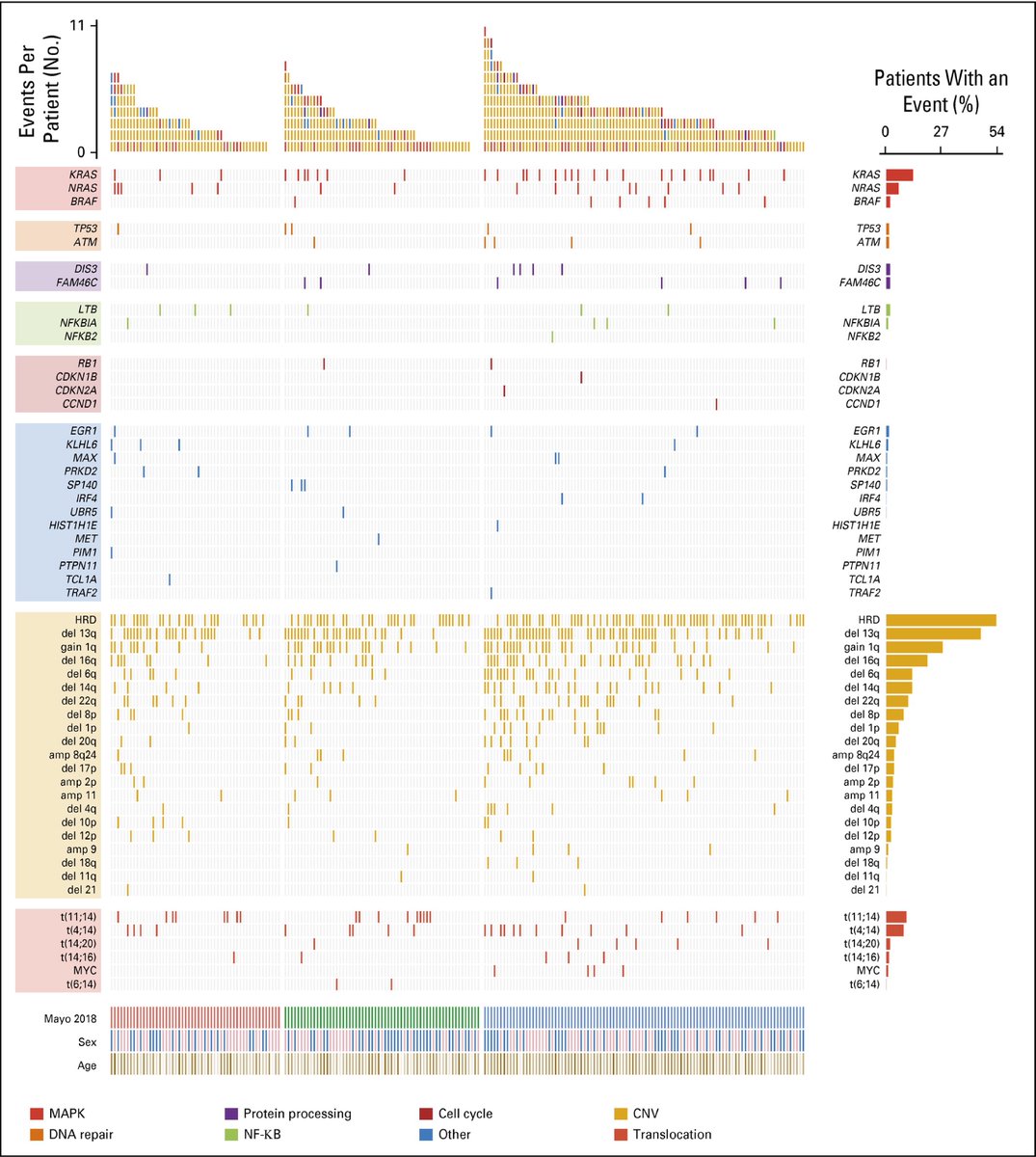
We profiled 214 diagnostic SMM tumor samples by exome(166) and targeted (48) sequencing. We found that the genomic landscape of SMM is the same as overt MM in terms of frequency and occurrence of all MM genomic events with exception to biallelic inactivation events
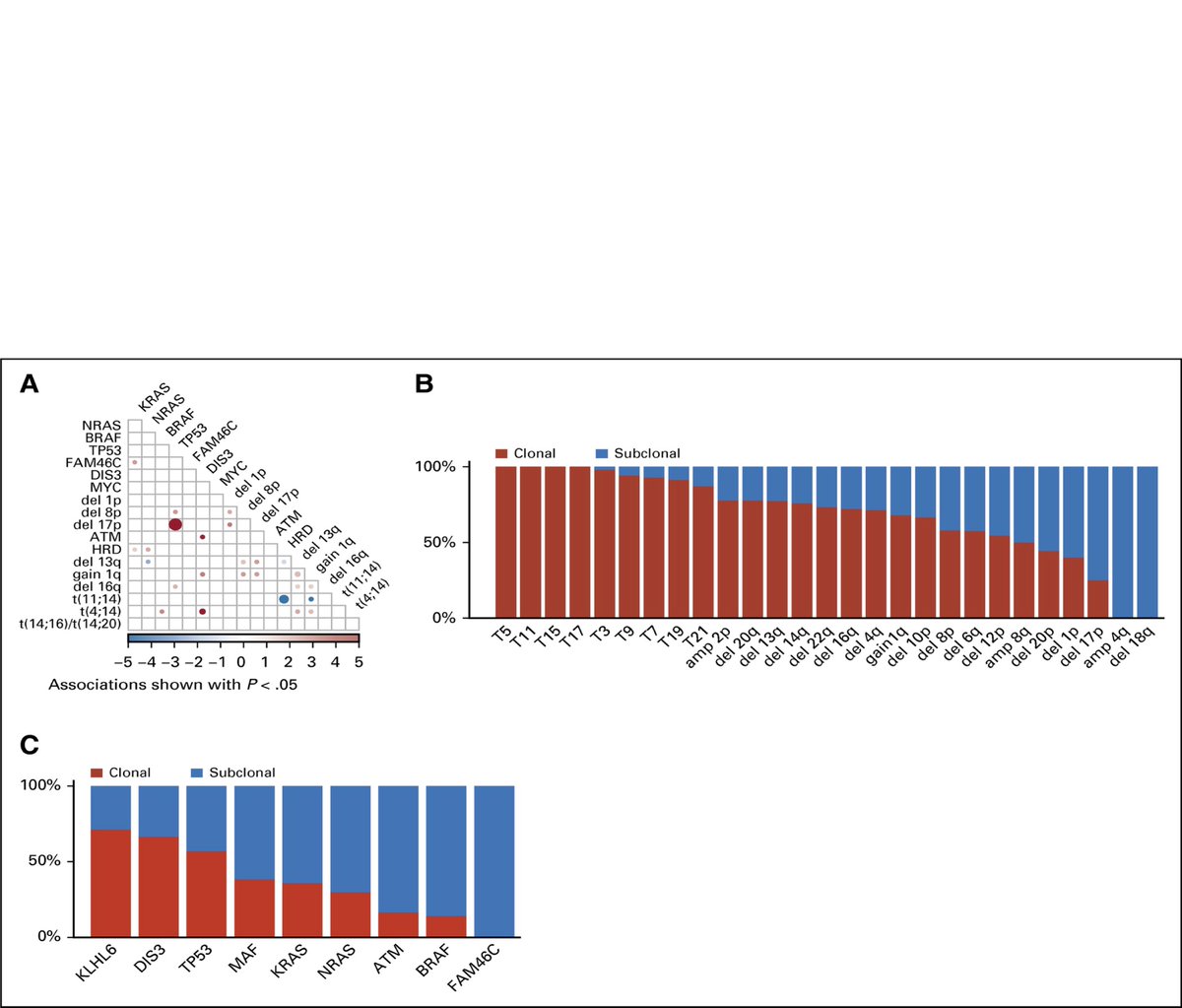
We have seen specific patterns of occurrence of different events. Some frequently occur together, while others are mutually exclusive. The clonality of CNAs and SNPs showed that CNAs are mostly clonal (earlier) events, while most of the SNPs are subclonal and occurring later.
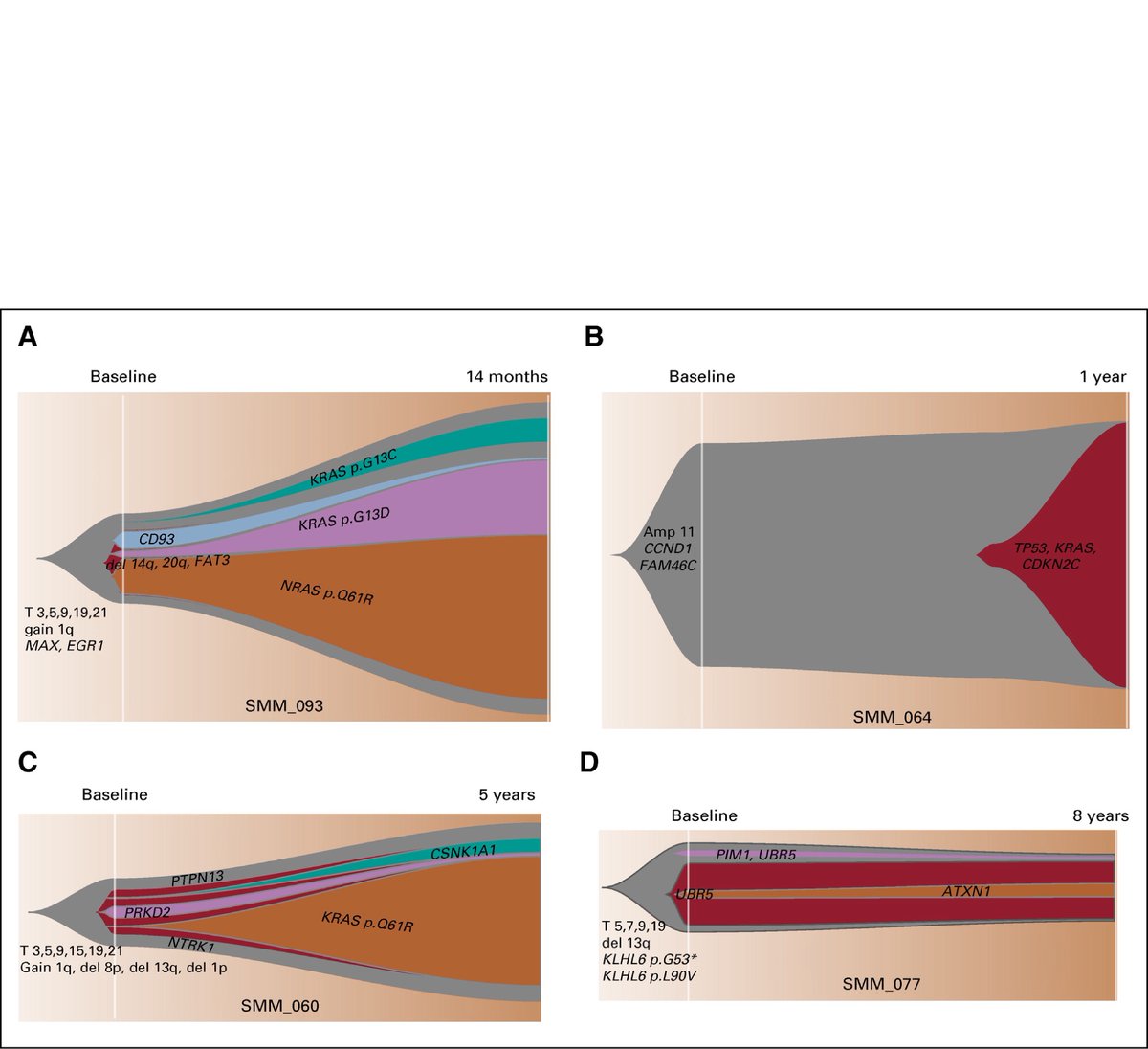
Also analyzing serial samples at time of diagnosis and time of progression or latest follow up (1-8 yrs) showed that most driver events were found at the SMM diagnosis, but with evidence of clonal evolution with subclonal CCFs changing over time and not static.
These findings indicate that what matters is the functional impact of these genomic alterations, which provide proliferative capacity to the tumor cells and within the microenvironment. So we looked at a multicenter cohort of 85 patients with a median follow up of 6.2 yrs.
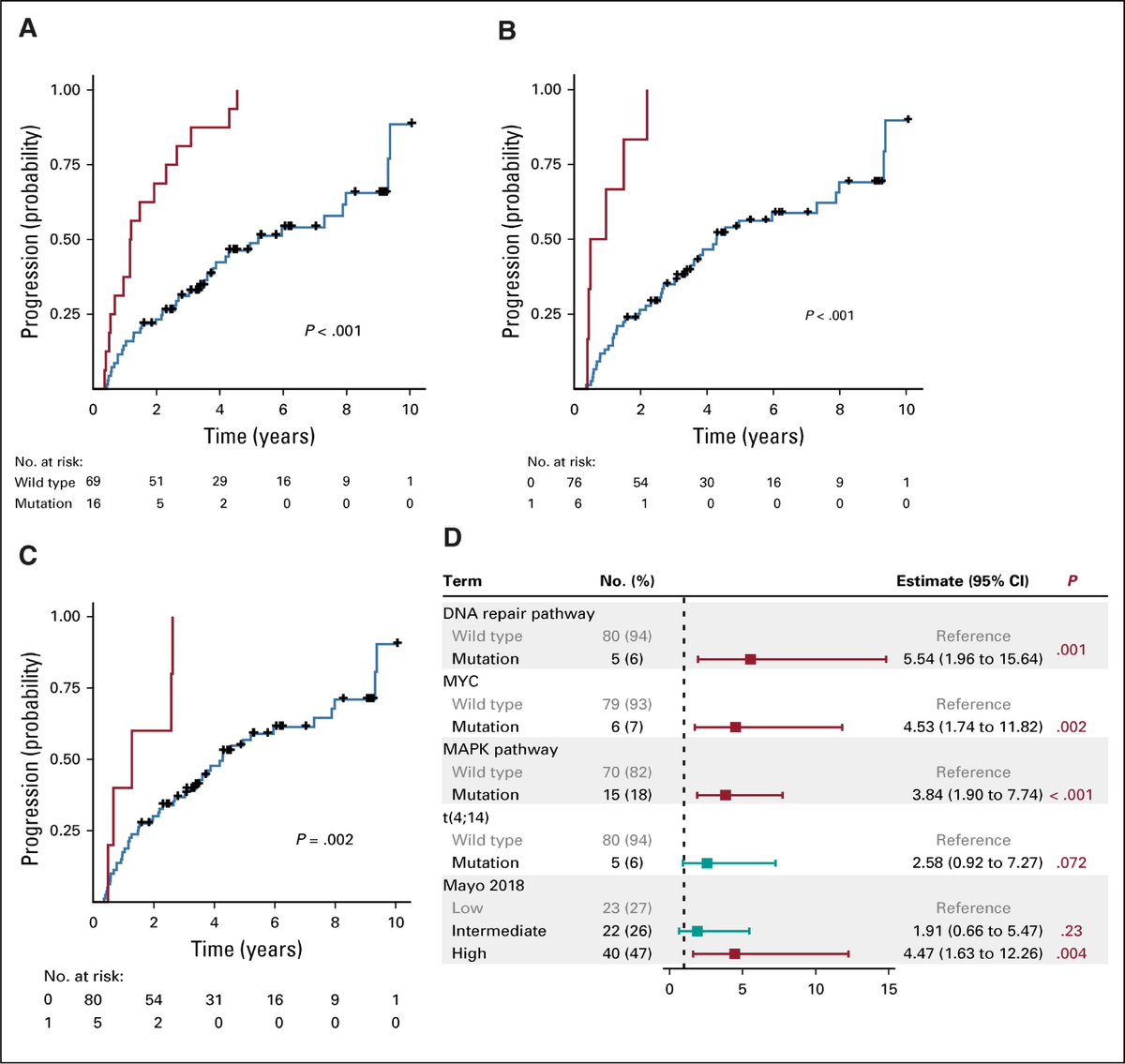
We wanted to identify biomarkers for progression that would predict high-risk SMM patients. We found that mutations in the MAPK and DNA repair pathways, as well as MYC translocations and amplifications are independent risk factors for progression to symptomatic MM.
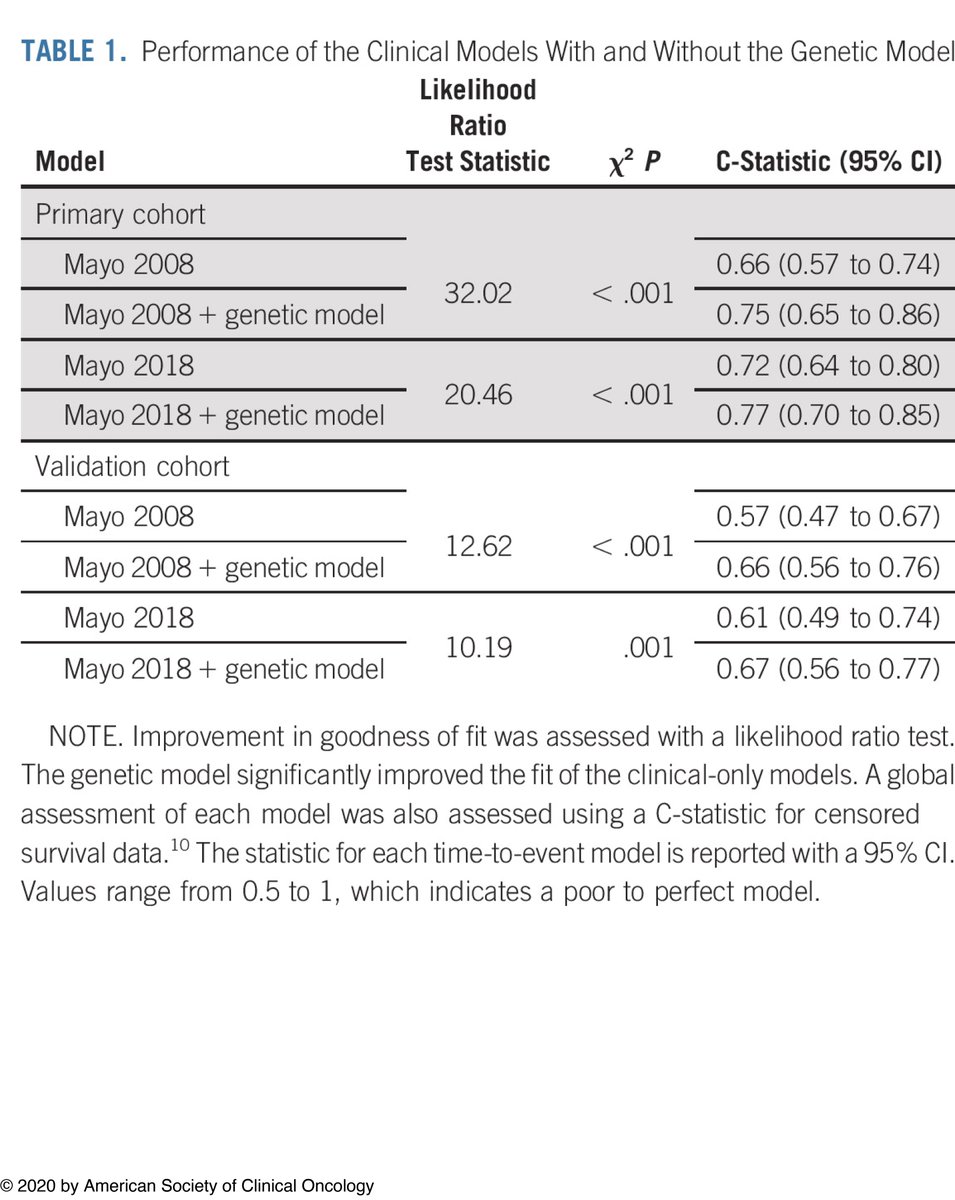
We developed a genomic model and tested it with the clinical risk models, such as Mayo 2018 and 2008. We found that the genomic model improves the prediction of progression more than clinical models alone. We used an external cohort from the Mayo Clinic to validate our results.
We found that using these high-risk genomic alterations significantly improve the prediction of progression in both our multi center and the validation cohorts, indicating that they can be used in risk stratification of SMM and identifying high-risk patients.
We found some other interesting biomarkers that were associated with a higher risk of progression on the univariate level only, such as t(4;14), APOBEC signature, and 1p and 8p deletions. These events could be a subject of larger studies to assess their impact.
This work could not have been possible without my dear colleague and study partner @RomanosSP and the support and guidance of our mentors @IreneGhobrial @PromiseStudy and @getz_lab @DanaFarber and @broadinstitute .
I would like to thank the whole study team and our collaborators from @uclcancer @thanosdimop @MayoClinic and @JanssenUS for their outstanding help and contribution. Also, many thanks to our funding sources @theNCI @theMMRF @SU2C @LLSResearch for their tremendous support

 Read on Twitter
Read on Twitter