Yesterday I ran a thread about our new paper using UK Biobank GWAS to identify core genes for several traits:
https://twitter.com/jkpritch/status/1253066314802839552
Today">https://twitter.com/jkpritch/... I want to expand on some fascinating things that we learned about sex differences in testosterone.
https://twitter.com/jkpritch/status/1253066314802839552
Today">https://twitter.com/jkpritch/... I want to expand on some fascinating things that we learned about sex differences in testosterone.
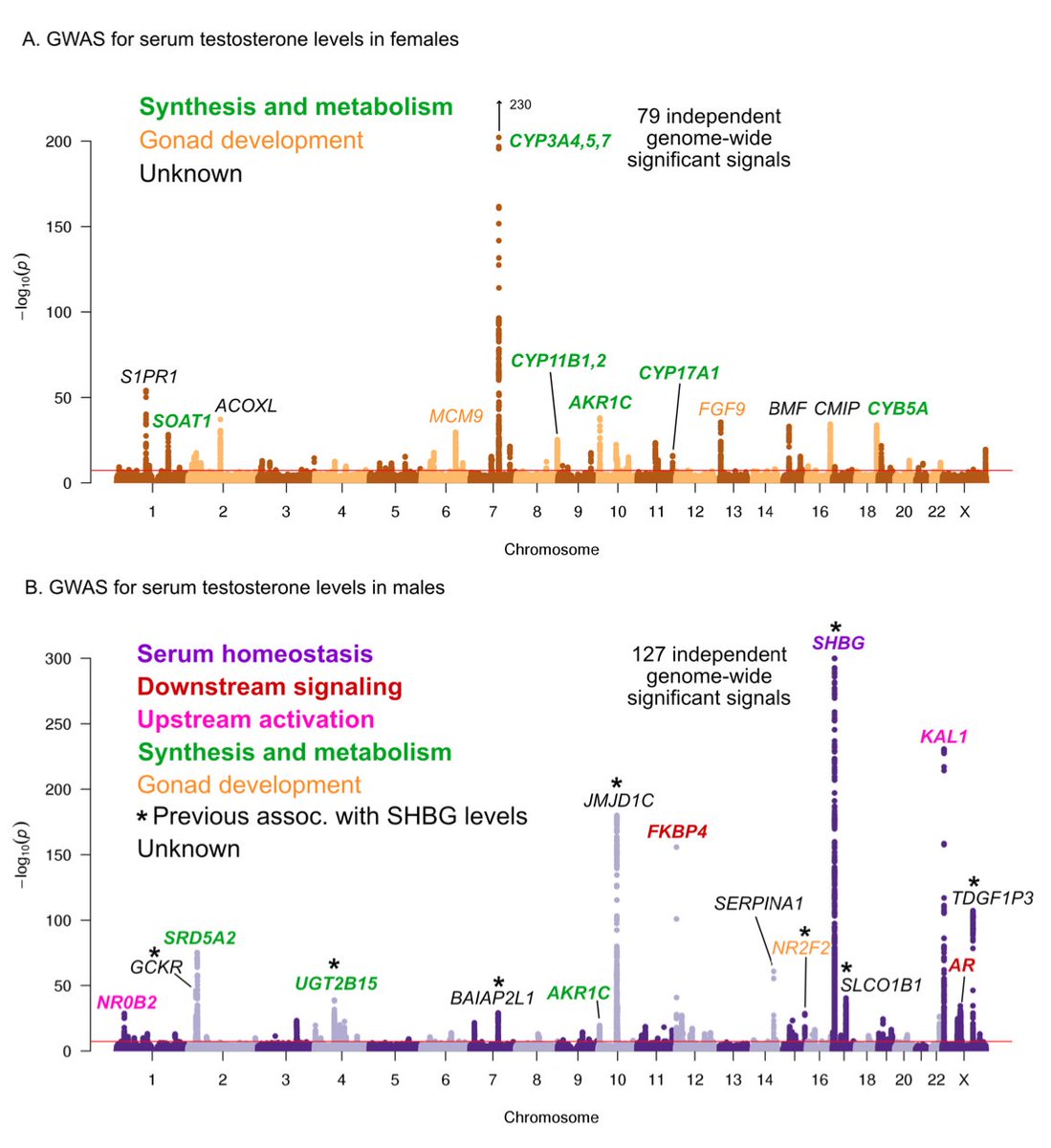
We and other groups have recently noticed strong sex differences between males and females in testosterone genetics (our analysis led by Sahin Naqvi @snaqvi1990 and Nasa Sinnott-Armstrong). Lead hits for testosterone share ~no overlap, and highlight different types of genes
Another way to look at this is by comparing effect sizes for hit SNPs in females vs males. No correlation. The insets show two other traits that are more typical (urate, SHBG). The genomewide genetic correlation by LDSR is also ~0.
Obviously, testosterone is produced at higher levels in males and females (~10x), and by different tissues (mainly testis in males; ovaries and adrenal in females). But to have ZERO correlation is very striking and unique as far as I know. Shouldn& #39;t some of the biology be shared?
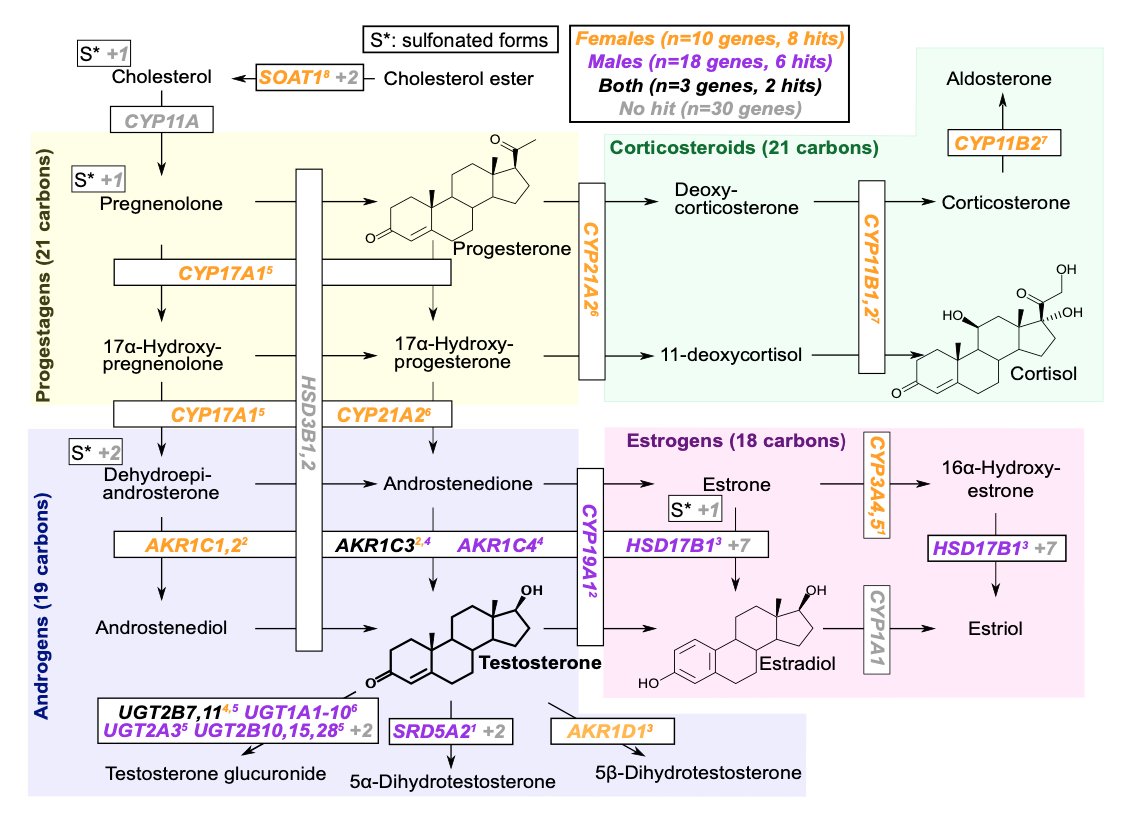
Testosterone is produced as one possible product of the steroid biosynthesis pathway. This is the most strongly enriched pathway for both sexes, but the genes show little overlap. Male hits mainly involve androgen processing, female hits progestagens.
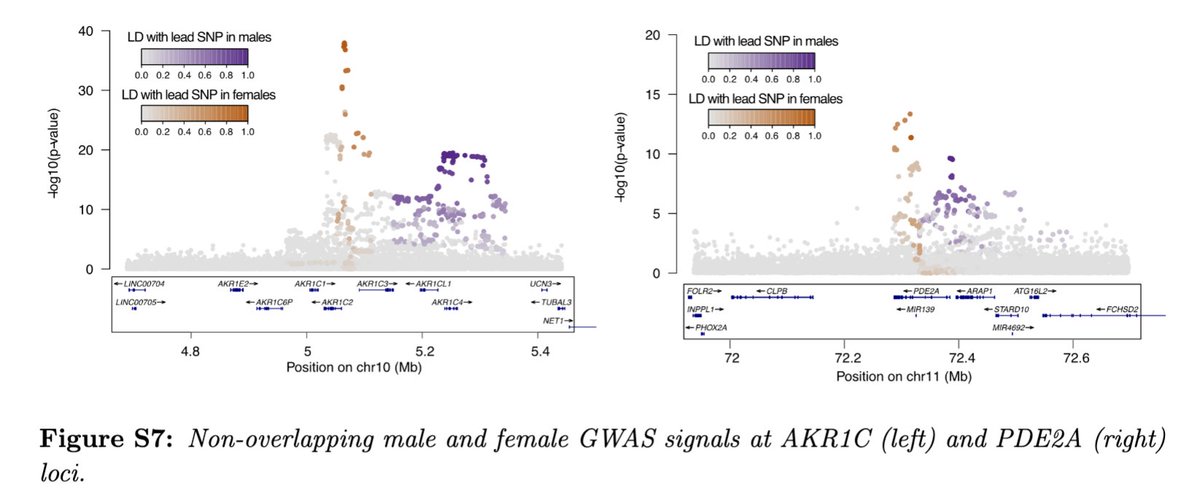
There are a couple of examples where both male and females have hits for biosynthetic enzymes in the same region but THESE DO NOT OVERLAP, presumably affecting different enhancers and/or paralogs (the mechanism is not clear yet).
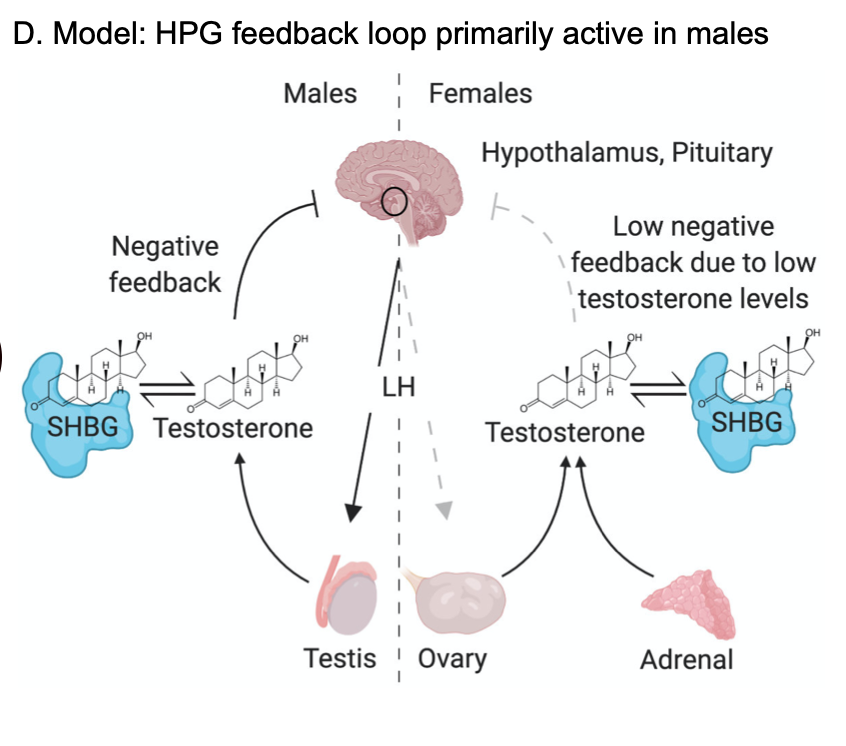
The next part of the story is that male testosterone is heavily regulated by the HPG (hypothalamic-pituitary-gonadal) axis.
We see numerous significant hits in the upstream signaling for males, but none in females. (CBAT= bioavailable T, more on that in a bit)
We see numerous significant hits in the upstream signaling for males, but none in females. (CBAT= bioavailable T, more on that in a bit)
The HPG axis seems to have little impact on female testosterone. This is partly because only ~half of female T comes from ovaries, but also because the impact of HPG on T in the ovaries is more complicated.
The last leg of the HPG axis is leuteinizing hormone (LH) signaling to the gonads. Consistent with the above, in males we see significant genetic correlation between LH and testosterone (0.27), but not in females (0.08). [Caveat is that the LH sample size is a bit low.]
There& #39;s one more key bit of this: active testosterone forms a negative feedback loop with the HPG, reducing T production. This is presumably why steroid users report testicular shrinkage (you can google for lurid stories about Barry Bonds, or less lurid
https://www.wwl.nhs.uk/News/2016/August/anabolic_steroids_effect_on_male_fertility.aspx">https://www.wwl.nhs.uk/News/2016...
https://www.wwl.nhs.uk/News/2016/August/anabolic_steroids_effect_on_male_fertility.aspx">https://www.wwl.nhs.uk/News/2016...
However, about half of all testosterone is bound by SHBG (steroid hormone binding globulin). The bound component is biologically unavailable and DOES NOT TRIGGER THE FEEDBACK. In males SHBG is the top hit in the genome; many other variants associated with SHBG also affect male T
Next: SHBG in males and females are highly correlated, but female SHBG does not affect female testosterone. This is because (1) the overall HPG effect on T is weaker in females, and (2) because females have too little T to stimulate the feedback effect.
This was shown by a study in the 1980s that manipulated female testosterone levels to see if it induced feedback in the HPG. They had to drive T levels all the way up to male levels to get feedback. (Are you wondering how did they get IRB for this?)
So our model is that biosynthesis is the single biggest factor in genetics of female testosterone levels. In males, there& #39;s a complicated dance between SHBG and HPG-axis function that winds up being much more important.

 Read on Twitter
Read on Twitter