I am certainly a big fan of using Mendelian disease genes to prioritize causal genes at GWAS loci so this tool seems like a step in the right direction. Still I am perplexed by the example selected to demonstrate the utility of the approach: ( @drpaternoster ) https://twitter.com/GWAS_lit/status/1252293521186467843">https://twitter.com/GWAS_lit/...
The trait is male pattern baldness and the SNP is rs7961185. There are 3 reliable heuristics for identifying the causal gene: 1) closest gene, 2) missense mutation, 3) relevant rare disease. In this case all 3 heuristics overlap in one gene: KRT71.
KRT71 is the closest gene using either TSS or TSS+TES: https://www.ncbi.nlm.nih.gov/snp/rs7961185 ,">https://www.ncbi.nlm.nih.gov/snp/rs796... https://genetics.opentargets.org/variant/12_52538808_G_T.
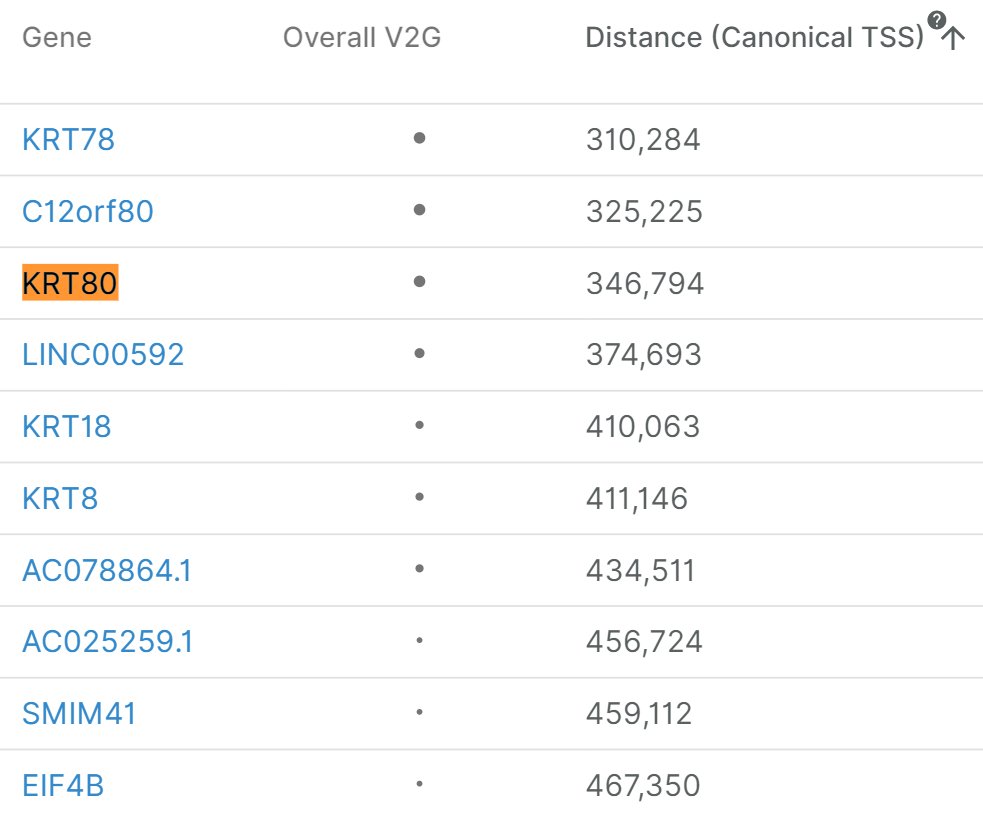
rs7961185">https://genetics.opentargets.org/variant/1... is in very high LD (r2=0.98) with a missense mutation in KRT71: https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php.">https://pubs.broadinstitute.org/mammals/h... According to @targetvalidate KRT80 is the 33rd closest gene at 350kb away
rs7961185">https://genetics.opentargets.org/variant/1... is in very high LD (r2=0.98) with a missense mutation in KRT71: https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php.">https://pubs.broadinstitute.org/mammals/h... According to @targetvalidate KRT80 is the 33rd closest gene at 350kb away
Finally, the specific rare disease associated with KRT71 is hypotrichiosis https://en.wiktionary.org/wiki/hypotrichosis">https://en.wiktionary.org/wiki/hypo... https://www.omim.org/entry/608245
In">https://www.omim.org/entry/608... fact I just learned that what makes the Sphinx cat hairless is a mutation in KRT71 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2974189/">https://www.ncbi.nlm.nih.gov/pmc/artic...
In">https://www.omim.org/entry/608... fact I just learned that what makes the Sphinx cat hairless is a mutation in KRT71 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2974189/">https://www.ncbi.nlm.nih.gov/pmc/artic...
So KRT71 would be a very reasonable suggestion for the causal gene. The presence of 19 paralogs at the locus clearly complicates things.
Bonus cat - turns out the Devon Rex breed is due to an alternate allele at KRT71
https://en.wikipedia.org/wiki/Devon_Rex ">https://en.wikipedia.org/wiki/Devo...
Bonus cat - turns out the Devon Rex breed is due to an alternate allele at KRT71
https://en.wikipedia.org/wiki/Devon_Rex ">https://en.wikipedia.org/wiki/Devo...

 Read on Twitter
Read on Twitter