So excited to finally share this work - structure of Cx46/50 gap junction intercellular channels in a dynamic lipid environment, resolved at 1.9 Å by #CryoEM! With some MD-simulation to make it dance ;) Check it out on BioRxiv: http://shorturl.at/iEW34 ">https://shorturl.at/iEW34&quo...
Lead by an extraordinary team of grad/undergrad students: Jonny Flores ( @CryoEMJonny), Bassam Haddad ( @BassBioPhys), Kim Dolan and Janette Myers – and our wonderful collaborators Craig Yoshioka ( @craigky), Jeremy Copperman and Dan Zuckerman @psu_research, @OHSUSOM
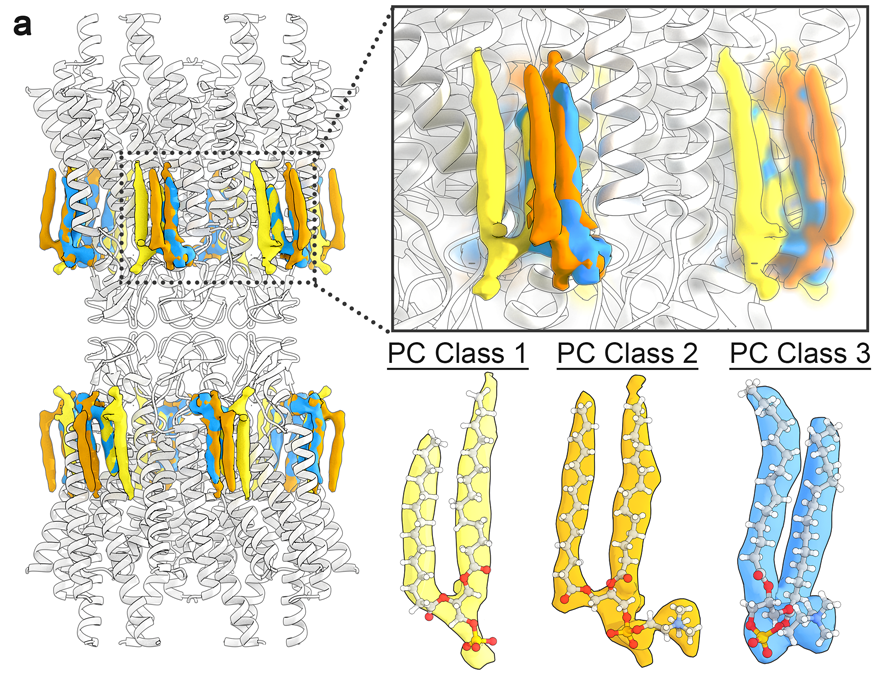
We see a stunning effect of Cx46/50 on the local lipid environment - effectively inducing a phase separation (to the gel state) that is specific to the extracellular lipid leaflet of the two opposed membranes
3D heterogeneity analysis of the CryoEM data identified multiple lipid configurations that co-exist within the dynamic lattice of stabilized lipids – and further detailed by MD
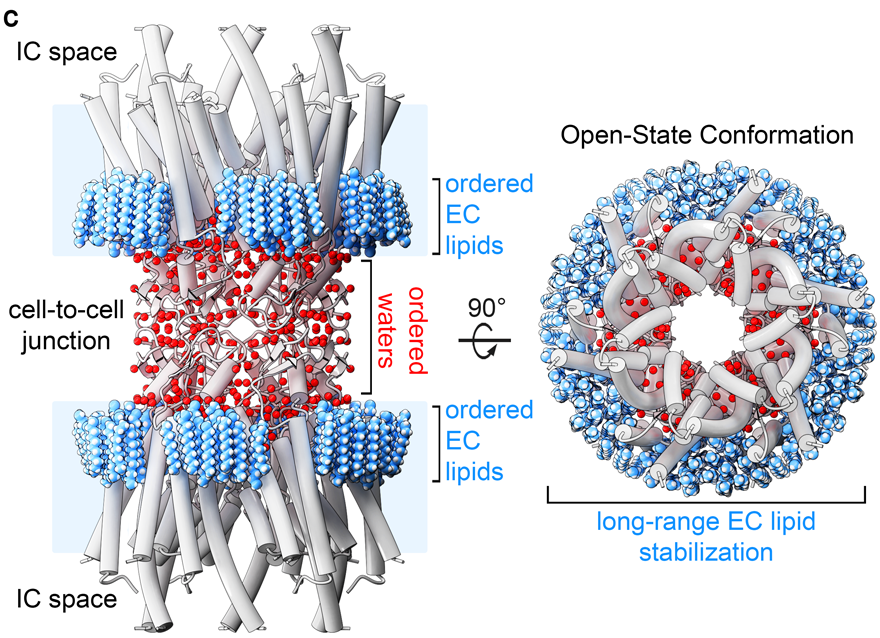
In addition, ~400 water molecules are resolved in the CryoEM map, localized at architectural and functionally important sites
Together this work uncovers previously unrecognized roles of the aqueous-lipid environment in stabilizing the structure and assembly of the gap junctions, and suggest Cx46/50 plays an important role in shaping the properties of local membrane environment
Thanks to @NIGMS and @CryoEM_PNCC for providing us the resources, and enabling us to have so much fun doing what we love! And to @SjorsScheres #Relion3.1 and @thermosciEMSpec #Krios #FalconIII for giving us the tools to do this work!

 Read on Twitter
Read on Twitter