There are a lot of phylogenetic trees  https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌳" title="Deciduous tree" aria-label="Emoji: Deciduous tree">getting bounced about in the midst of #COVID19 and they’re getting really really big. A phylogenetic tree is a valuable resource mid #pandemic but they& #39;re not always easy to interpret.
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌳" title="Deciduous tree" aria-label="Emoji: Deciduous tree">getting bounced about in the midst of #COVID19 and they’re getting really really big. A phylogenetic tree is a valuable resource mid #pandemic but they& #39;re not always easy to interpret.
Here& #39;s a brief explainer on just one aspect. 1/11
Here& #39;s a brief explainer on just one aspect. 1/11
A https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌳" title="Deciduous tree" aria-label="Emoji: Deciduous tree"> reconstructs the evolutionary relationships between viruses; those which are more closely related to each other and those which are more distant. We build these trees by comparing
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌳" title="Deciduous tree" aria-label="Emoji: Deciduous tree"> reconstructs the evolutionary relationships between viruses; those which are more closely related to each other and those which are more distant. We build these trees by comparing  https://abs.twimg.com/emoji/v2/... draggable="false" alt="🧬" title="DNA" aria-label="Emoji: DNA">s.
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🧬" title="DNA" aria-label="Emoji: DNA">s.
For #SARSCoV2 there are ~30,000 positions to look at but only a small fraction vary. 2/11
For #SARSCoV2 there are ~30,000 positions to look at but only a small fraction vary. 2/11
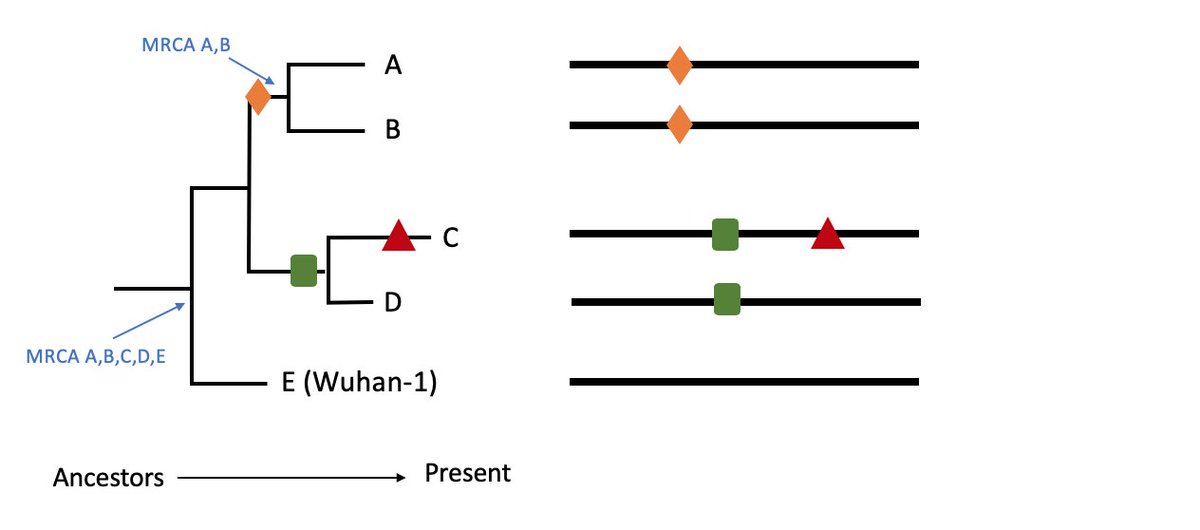
Let& #39;s take a toy example of five #SARSCov2.
This https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌳" title="Deciduous tree" aria-label="Emoji: Deciduous tree"> for example places A & B as more closely related to each other than to C, D or E (outgroup).
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌳" title="Deciduous tree" aria-label="Emoji: Deciduous tree"> for example places A & B as more closely related to each other than to C, D or E (outgroup).
Each tree node therefore provides the theoretical Most Recent Common Ancestor (MRCA) relative to the samples in the dataset. 3/11
This
Each tree node therefore provides the theoretical Most Recent Common Ancestor (MRCA) relative to the samples in the dataset. 3/11
The data underlying trees are the nucleotide changes (mutations - here coloured shapes) identified across the ~30,000 length sequence alignment.
Eg. here closely related A & B share the same orange diamond change (mutation) not seen in C, D or E. 4/11
Eg. here closely related A & B share the same orange diamond change (mutation) not seen in C, D or E. 4/11
These mutations are not necessarily a bad thing. They largely represent the gradual accumulation of, mainly harmless, typos which can occur during replication
It is these mutations that allow us to log & trace the pandemic using https://abs.twimg.com/emoji/v2/... draggable="false" alt="🧬" title="DNA" aria-label="Emoji: DNA"> data.
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🧬" title="DNA" aria-label="Emoji: DNA"> data.
More here: http://tinyurl.com/vs9gqxg ">https://tinyurl.com/vs9gqxg&q... 5/11
It is these mutations that allow us to log & trace the pandemic using
More here: http://tinyurl.com/vs9gqxg ">https://tinyurl.com/vs9gqxg&q... 5/11
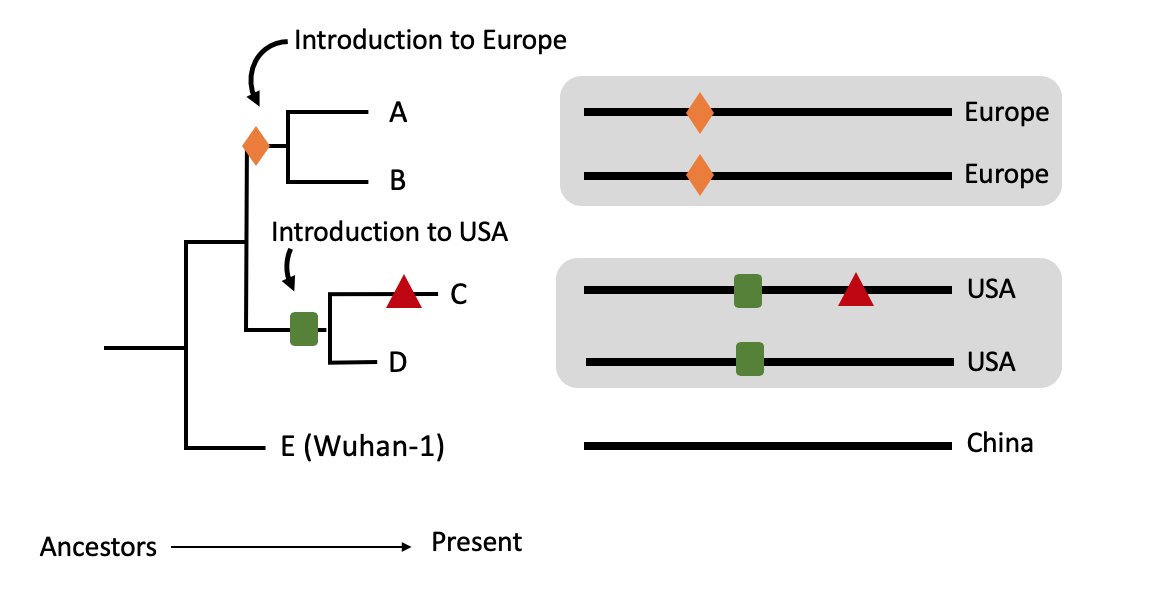
In addition for each https://abs.twimg.com/emoji/v2/... draggable="false" alt="🧬" title="DNA" aria-label="Emoji: DNA">we know where in the world it was sampled.
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🧬" title="DNA" aria-label="Emoji: DNA">we know where in the world it was sampled.
Say A & B are from Europe, C & D from the USA & E from China.
We can infer a likely scenario whereby the ancestor of A & B (MRCA A,B) was in Europe and the ancestor of C & D (MRCA C,D) was in the USA. 6/11
Say A & B are from Europe, C & D from the USA & E from China.
We can infer a likely scenario whereby the ancestor of A & B (MRCA A,B) was in Europe and the ancestor of C & D (MRCA C,D) was in the USA. 6/11
Though note the above inference is always relative to what we have sequenced.
The available genetic data is getting large BUT it is still only a very small portion of the diversity circulating globally https://abs.twimg.com/emoji/v2/... draggable="false" alt="🗺️" title="World map" aria-label="Emoji: World map">
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🗺️" title="World map" aria-label="Emoji: World map">
...though this may be the most densely sequenced outbreak to date. 7/11
The available genetic data is getting large BUT it is still only a very small portion of the diversity circulating globally
...though this may be the most densely sequenced outbreak to date. 7/11
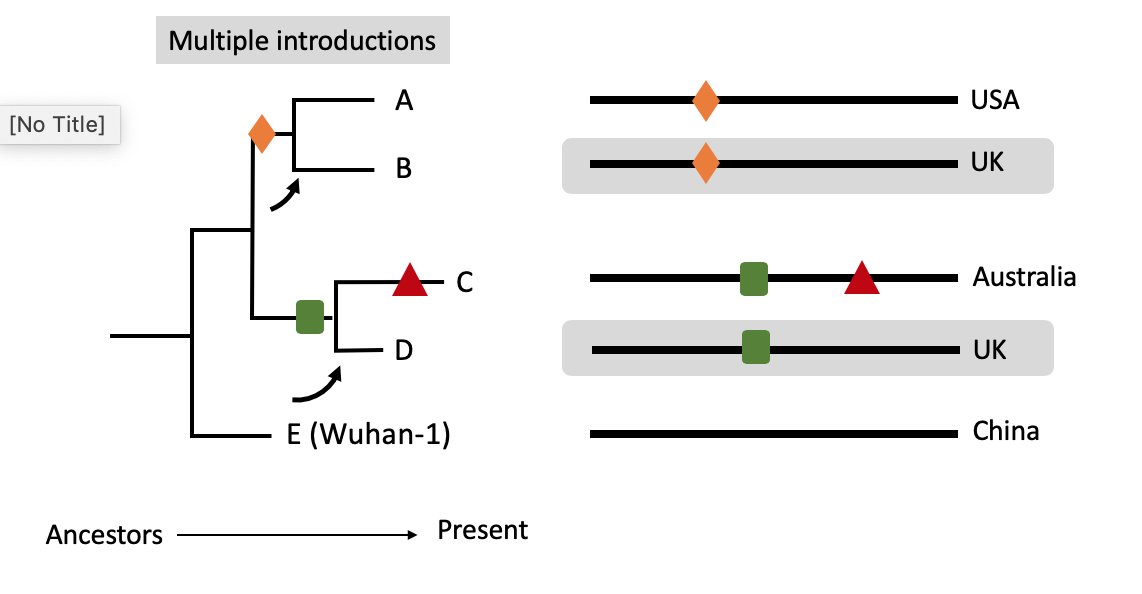
For viruses sampled in the UK we see related clades falling in multiple locations in the https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌳" title="Deciduous tree" aria-label="Emoji: Deciduous tree">& interspersed with those from other regions.
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌳" title="Deciduous tree" aria-label="Emoji: Deciduous tree">& interspersed with those from other regions.
Eg. here the location of B & D suggests >1 UK introduction.
These can then seed local transmissions leading to expanding related clades. 8/11
Eg. here the location of B & D suggests >1 UK introduction.
These can then seed local transmissions leading to expanding related clades. 8/11
Here is the actual (WIP) #SARSCoV2  https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌳" title="Deciduous tree" aria-label="Emoji: Deciduous tree">highlighting the multiple placements of UK viruses: https://twitter.com/alanmcn1/status/1249656885969661952">https://twitter.com/alanmcn1/...
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌳" title="Deciduous tree" aria-label="Emoji: Deciduous tree">highlighting the multiple placements of UK viruses: https://twitter.com/alanmcn1/status/1249656885969661952">https://twitter.com/alanmcn1/...
(Much prettier trees are available as queryable visualisations on the wonderful @nextstrain aided by huge volumes of data shared by the community on @GISAID.)
9/11
(Much prettier trees are available as queryable visualisations on the wonderful @nextstrain aided by huge volumes of data shared by the community on @GISAID.)
9/11
But the UK is not unusual. Similar patterns are seen for many densely sequenced countries.
The new “Regional” tab in @nextstrain provides an easy way to view these.
Eg. see https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Down pointing backhand index" aria-label="Emoji: Down pointing backhand index"> for viruses sampled in the USA (enlarged light green tips) falling all across the global tree. 10/11
https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Down pointing backhand index" aria-label="Emoji: Down pointing backhand index"> for viruses sampled in the USA (enlarged light green tips) falling all across the global tree. 10/11
The new “Regional” tab in @nextstrain provides an easy way to view these.
Eg. see
Phylogenetic trees provide powerful surveillance tools pointing to multiple introductions of #SARSCoV2 to countries all around the  https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌍" title="Earth globe europe-africa" aria-label="Emoji: Earth globe europe-africa">followed by local #transmissions.
https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌍" title="Earth globe europe-africa" aria-label="Emoji: Earth globe europe-africa">followed by local #transmissions.
For more on inference from phylogenies see this brilliant webinar by @firefoxx66: https://tinyurl.com/v9xpucj .">https://tinyurl.com/v9xpucj&q... 11/11
For more on inference from phylogenies see this brilliant webinar by @firefoxx66: https://tinyurl.com/v9xpucj .">https://tinyurl.com/v9xpucj&q... 11/11

 Read on Twitter
Read on Twitter for example places A & B as more closely related to each other than to C, D or E (outgroup).Each tree node therefore provides the theoretical Most Recent Common Ancestor (MRCA) relative to the samples in the dataset. 3/11" title="Let& #39;s take a toy example of five #SARSCov2. This https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌳" title="Deciduous tree" aria-label="Emoji: Deciduous tree"> for example places A & B as more closely related to each other than to C, D or E (outgroup).Each tree node therefore provides the theoretical Most Recent Common Ancestor (MRCA) relative to the samples in the dataset. 3/11" class="img-responsive" style="max-width:100%;"/>
for example places A & B as more closely related to each other than to C, D or E (outgroup).Each tree node therefore provides the theoretical Most Recent Common Ancestor (MRCA) relative to the samples in the dataset. 3/11" title="Let& #39;s take a toy example of five #SARSCov2. This https://abs.twimg.com/emoji/v2/... draggable="false" alt="🌳" title="Deciduous tree" aria-label="Emoji: Deciduous tree"> for example places A & B as more closely related to each other than to C, D or E (outgroup).Each tree node therefore provides the theoretical Most Recent Common Ancestor (MRCA) relative to the samples in the dataset. 3/11" class="img-responsive" style="max-width:100%;"/>
 we know where in the world it was sampled.Say A & B are from Europe, C & D from the USA & E from China.We can infer a likely scenario whereby the ancestor of A & B (MRCA A,B) was in Europe and the ancestor of C & D (MRCA C,D) was in the USA. 6/11" title="In addition for eachhttps://abs.twimg.com/emoji/v2/... draggable="false" alt="🧬" title="DNA" aria-label="Emoji: DNA">we know where in the world it was sampled.Say A & B are from Europe, C & D from the USA & E from China.We can infer a likely scenario whereby the ancestor of A & B (MRCA A,B) was in Europe and the ancestor of C & D (MRCA C,D) was in the USA. 6/11" class="img-responsive" style="max-width:100%;"/>
we know where in the world it was sampled.Say A & B are from Europe, C & D from the USA & E from China.We can infer a likely scenario whereby the ancestor of A & B (MRCA A,B) was in Europe and the ancestor of C & D (MRCA C,D) was in the USA. 6/11" title="In addition for eachhttps://abs.twimg.com/emoji/v2/... draggable="false" alt="🧬" title="DNA" aria-label="Emoji: DNA">we know where in the world it was sampled.Say A & B are from Europe, C & D from the USA & E from China.We can infer a likely scenario whereby the ancestor of A & B (MRCA A,B) was in Europe and the ancestor of C & D (MRCA C,D) was in the USA. 6/11" class="img-responsive" style="max-width:100%;"/>
 & interspersed with those from other regions.Eg. here the location of B & D suggests >1 UK introduction. These can then seed local transmissions leading to expanding related clades. 8/11" title="For viruses sampled in the UK we see related clades falling in multiple locations in thehttps://abs.twimg.com/emoji/v2/... draggable="false" alt="🌳" title="Deciduous tree" aria-label="Emoji: Deciduous tree">& interspersed with those from other regions.Eg. here the location of B & D suggests >1 UK introduction. These can then seed local transmissions leading to expanding related clades. 8/11" class="img-responsive" style="max-width:100%;"/>
& interspersed with those from other regions.Eg. here the location of B & D suggests >1 UK introduction. These can then seed local transmissions leading to expanding related clades. 8/11" title="For viruses sampled in the UK we see related clades falling in multiple locations in thehttps://abs.twimg.com/emoji/v2/... draggable="false" alt="🌳" title="Deciduous tree" aria-label="Emoji: Deciduous tree">& interspersed with those from other regions.Eg. here the location of B & D suggests >1 UK introduction. These can then seed local transmissions leading to expanding related clades. 8/11" class="img-responsive" style="max-width:100%;"/>
 for viruses sampled in the USA (enlarged light green tips) falling all across the global tree. 10/11" title="But the UK is not unusual. Similar patterns are seen for many densely sequenced countries.The new “Regional” tab in @nextstrain provides an easy way to view these. Eg. see https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Down pointing backhand index" aria-label="Emoji: Down pointing backhand index"> for viruses sampled in the USA (enlarged light green tips) falling all across the global tree. 10/11" class="img-responsive" style="max-width:100%;"/>
for viruses sampled in the USA (enlarged light green tips) falling all across the global tree. 10/11" title="But the UK is not unusual. Similar patterns are seen for many densely sequenced countries.The new “Regional” tab in @nextstrain provides an easy way to view these. Eg. see https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Down pointing backhand index" aria-label="Emoji: Down pointing backhand index"> for viruses sampled in the USA (enlarged light green tips) falling all across the global tree. 10/11" class="img-responsive" style="max-width:100%;"/>


