Kinases are often tethered to their substrates via disordered linkers or scaffolding proteins. But how does the properties of the linker affect phosphorylation? That is what @dylamateusz and I are trying to answer in our new pre-print: https://www.biorxiv.org/content/10.1101/2020.04.04.023713v1">https://www.biorxiv.org/content/1...
Kinases often have a switch-like roles, where they only phosphorylate a few substrates to exert their effect. The specificity of such reactions is maintain by physically connecting the enzyme to the substrate typically via a disordered protein/region.
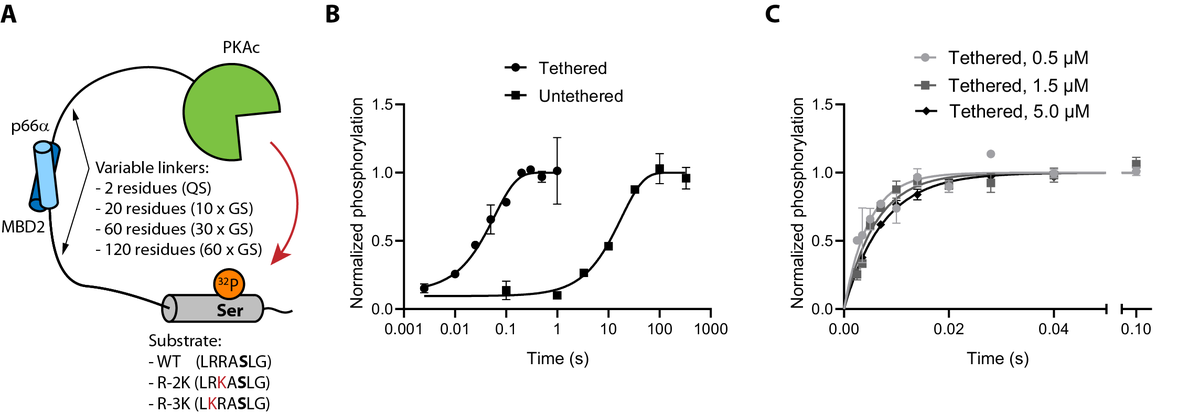
Here, we investigate to the role of the linker in intra-complex phosphorylation reactions. Tethered reactions are fundamentally different from Michaelis-Menten like reactions as they are effectively single turnover.
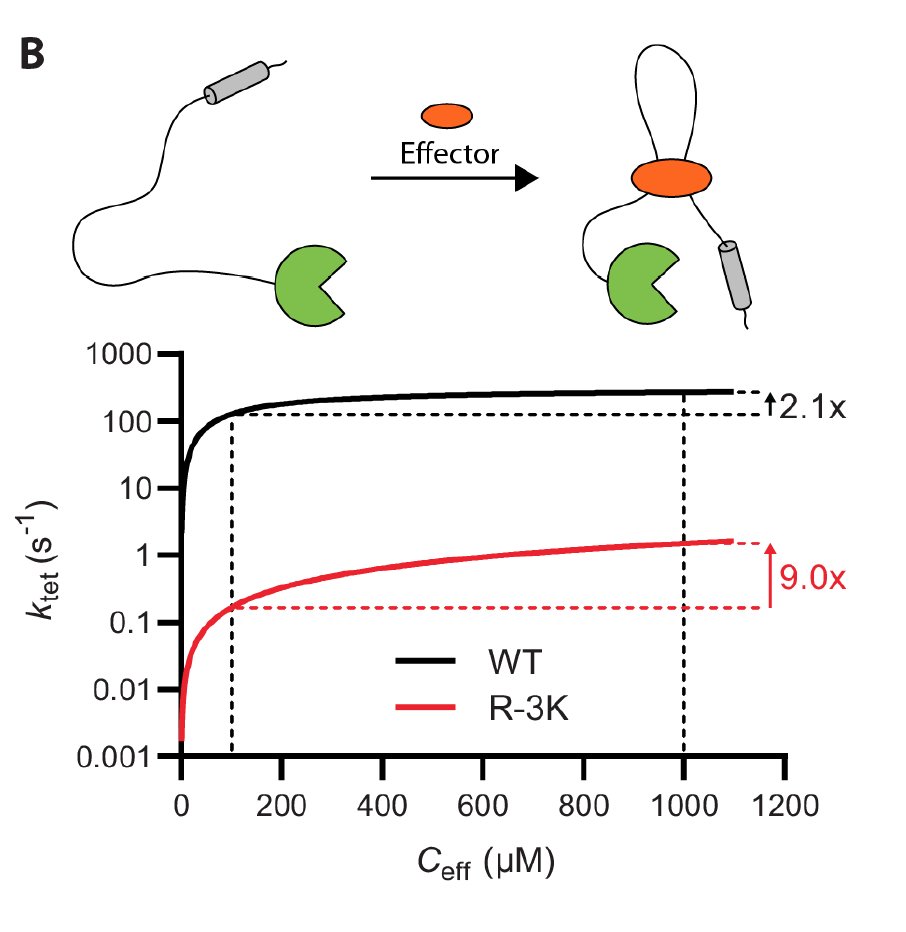
We made a model system consisting of a kinase tethered to its substrate via disordered linkers, and measure the rate of intra-molecular phosphorylation. Tethering accelerates the reaction by ~100-fold, but is independent of concentration.
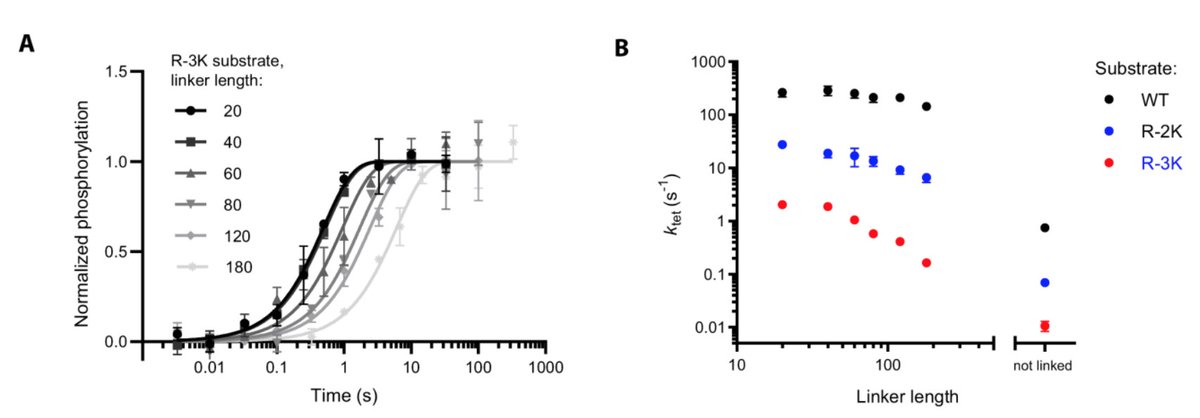
Phosphorylation rates scale with the length of the linker (longer = slower), but the magnitude of the linker scaling depends on the substrate. For the same linker variants we see a 2-fold change for one substrate, and a 10-fold for another.
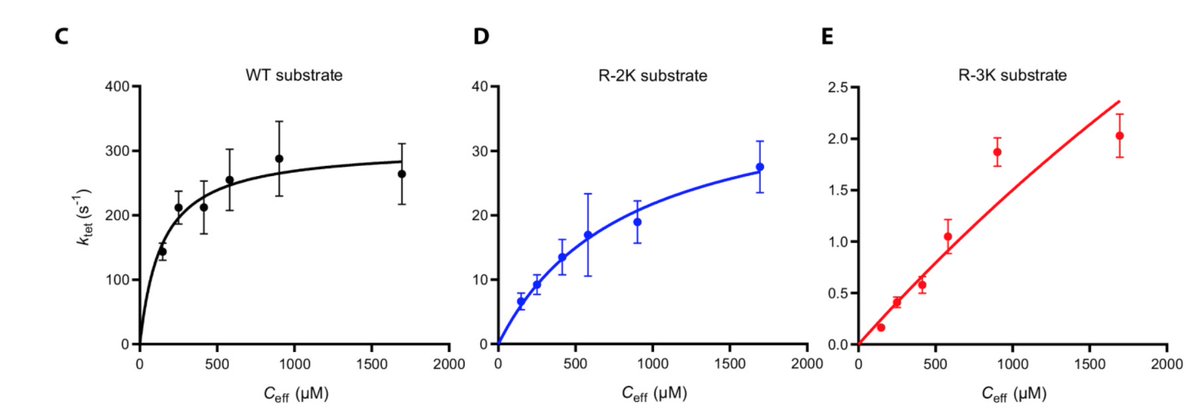
We find that the reaction rate can be described by a Michaelis-Menten like dependence on the effective concentration enforced by the linker. This means that the reaction saturates at high effective concentrations and will increase if the linker is shortened.
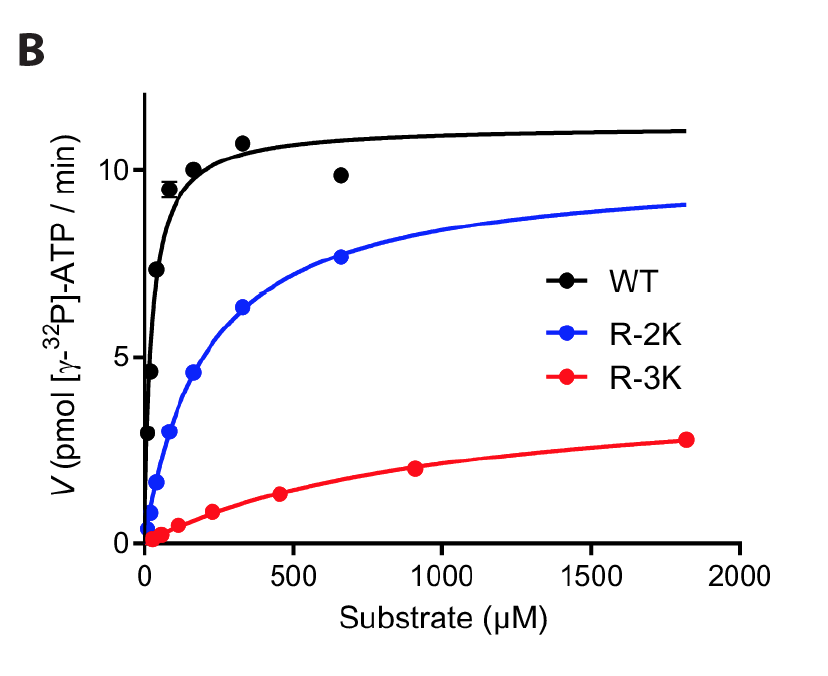
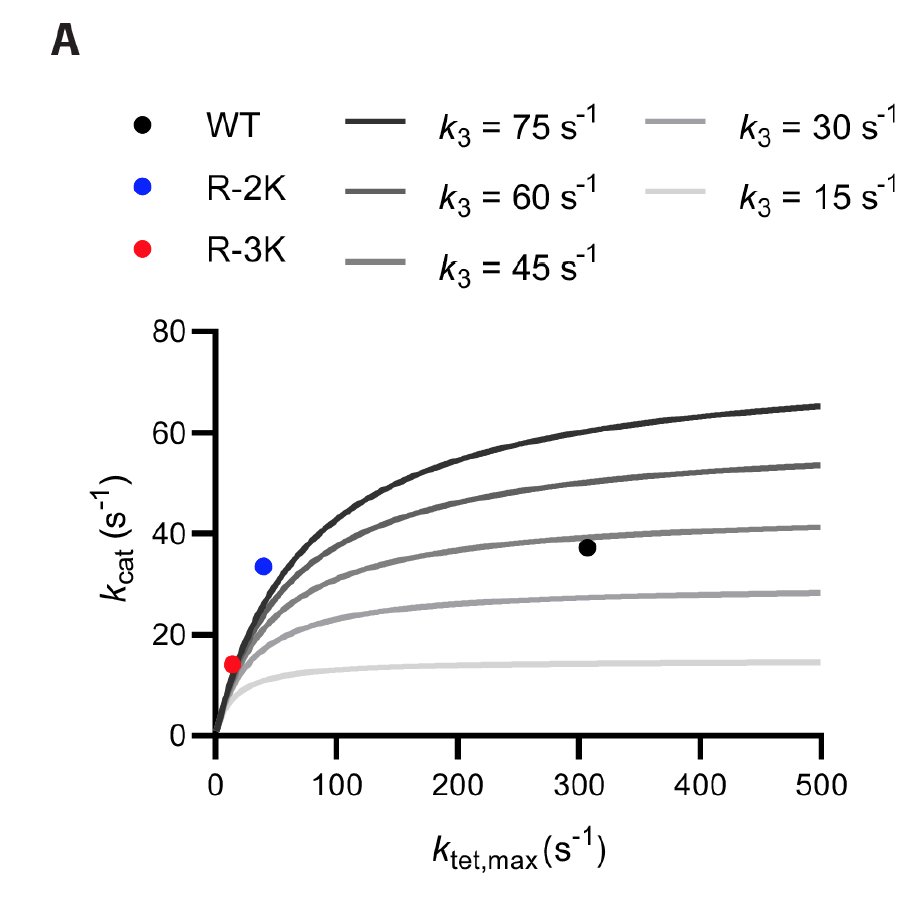
Steady-state kinetic parameters only have a limited ability to predict tethered rates: We have variants with 8-fold different maximal tethered rates, but similar k(cat).
This occurs because steady-state phosphorylation reactions are mostly limited by product release, which may disguise big differences in how quickly substrates are phosphorylated in a tethered system.
We suggest how changes in linker regions can allosterically shift signalling output: As substrates saturate at different effective concentrations, an increase in the effective concentration shift the relative substrate usage towards low affinity substrates.
Also, if you like this thread, you may also want to check out a recent pre-print from the group Jesse Zalatan: Using a similar design, they get rates that are ~10.000-fold lower. We do not know the reason for this difference, but suggestions are welcome: https://www.biorxiv.org/content/10.1101/2020.03.12.989012v1">https://www.biorxiv.org/content/1...
And finally a big thanks to @Villumfonden for funding this project, although the proposal was admittedly quite abstract and hand-wavy and the applicant didn’t have any preliminary data.

 Read on Twitter
Read on Twitter