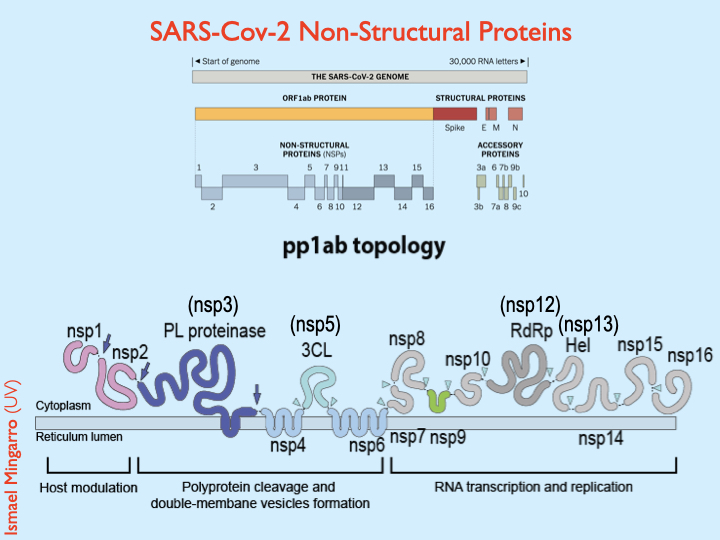
#SARSCoV2 Proteome includes 29 proteins: 16 non-structural proteins + 4 structural proteins + 9 accessory proteins. This scheme from @ViralZone @ISBSIB shows where are they encoded within the viral RNA genome
#TPbiotecUV #BqCBemeUV Thread https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Down pointing backhand index" aria-label="Emoji: Down pointing backhand index"> .1/7
https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Down pointing backhand index" aria-label="Emoji: Down pointing backhand index"> .1/7
#TPbiotecUV #BqCBemeUV Thread
#SARSCoV2 Non-Structural Proteins (NSPs) are synthesized as two large polyproteins pp1a and pp1ab that are processed by two viral endoproteases NSP5 and NSP3. In the scheme (bottom) you can see the membrane topology of the polyprotein1ab (pp1ab) and the digestion sites 2/7
#SARSCoV2 the 15 Non-Structural Proteins encoded in the pp1ab illustrated by @13pt and @carlzimmer at @NYTScience include the 2 proteases, the replicase, an helicase,...
Notice that NPS11 is not represented because it& #39;s only encoded in polyprotein1a 3/7
Notice that NPS11 is not represented because it& #39;s only encoded in polyprotein1a 3/7
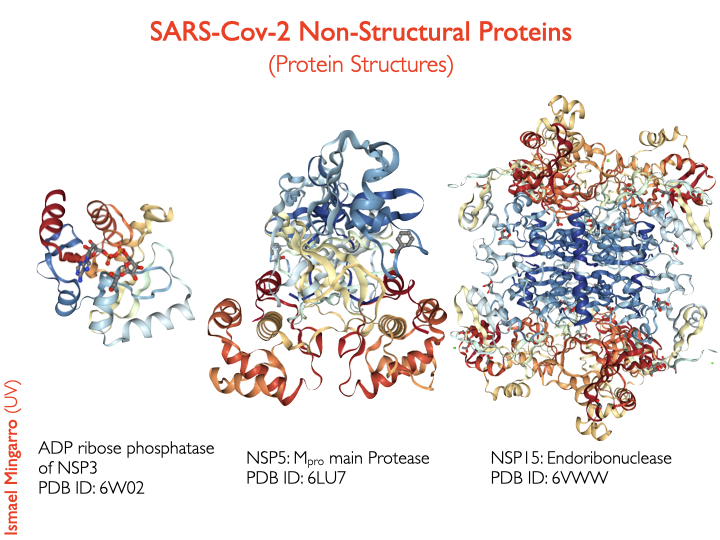
#SARSCoV2 the structures recently solved for some of this proteins are available at PDB-101 @buildmodels
Main protease structure is solved in complex with an inhibitor N3, which can be used as scafold to design new inhibitors
4/7
Main protease structure is solved in complex with an inhibitor N3, which can be used as scafold to design new inhibitors
4/7
#SARSCoV2 Structural Proteins: S (spike) and E (envelope) are single-spanning membrane proteins. S protein interacts with host ACE2 receptor. M (membrane) is a multispanning (likely 4TMs) membrane protein. N (nucleocapside) protects the RNA, keeping it stable inside the virus
5/7
5/7
#SARSCoV2 Structural proteins number of copies per virion have been estimated https://twitter.com/intent/tweet/?text=SARS-CoV-2%20%28COVID-19%29%20by%20the%20numbers&url=https%3A%2F%2Fdoi.org%2F10.7554%2FeLife.57309:
M:">https://twitter.com/intent/tw... 2000
N: 1000
S: 300 (arranged in 100 trimers)
E: 20
We have several structures for S protein (some with the receptor) and the RNA binding domain of N 6/7
M:">https://twitter.com/intent/tw... 2000
N: 1000
S: 300 (arranged in 100 trimers)
E: 20
We have several structures for S protein (some with the receptor) and the RNA binding domain of N 6/7
#SARSCoV2 Accessory proteins (AP) help change the environment inside the infected cell to make it easier for the virus to replicate @13pt @carlzimmer.
Proteins (5 from 9 AP) modeled by Yang Zhang’s Research Group @UMich 7/7
Proteins (5 from 9 AP) modeled by Yang Zhang’s Research Group @UMich 7/7

 Read on Twitter
Read on Twitter .1/7" title=" #SARSCoV2 Proteome includes 29 proteins: 16 non-structural proteins + 4 structural proteins + 9 accessory proteins. This scheme from @ViralZone @ISBSIB shows where are they encoded within the viral RNA genome #TPbiotecUV #BqCBemeUV Thread https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Down pointing backhand index" aria-label="Emoji: Down pointing backhand index"> .1/7" class="img-responsive" style="max-width:100%;"/>
.1/7" title=" #SARSCoV2 Proteome includes 29 proteins: 16 non-structural proteins + 4 structural proteins + 9 accessory proteins. This scheme from @ViralZone @ISBSIB shows where are they encoded within the viral RNA genome #TPbiotecUV #BqCBemeUV Thread https://abs.twimg.com/emoji/v2/... draggable="false" alt="👇" title="Down pointing backhand index" aria-label="Emoji: Down pointing backhand index"> .1/7" class="img-responsive" style="max-width:100%;"/>